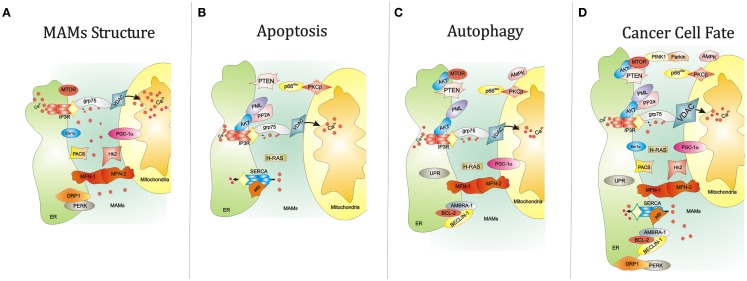
Figure 1.
Summary of the key functions of the ER-MAMs resident proteins. (A) Several proteins reside in MAMs compartment and regulate the juxtaposition between ER and mitochondria, including grp75, Ero1α, PERK, and MFN-1/2. At the same time, other MAM resident proteins control the cell survival by governing apoptosis (B) and autophagy (C). For example, it has been reported that PML, IP3R3, and AKT mutually interact to allow the correct Ca2+-movement between ER and mitochondria, an essential proapoptotic signal. Interestingly, the maintenance of this interorganelle Ca2+-communication is also important for the autophagic process. Of relevance, reductions of mitochondrial Ca2+ accumulation may trigger autophagy. In parallel, most of MAM proteins also govern the autophagic machinery. Of relevance, AKT and MTOR regulate negatively autophagic process. The same work is executed by the molecular axis composed by p66Shc and PKCβ. It is widely accepted that autophagy is strictly linked to several human diseases, in particular cancer. Think about a possible link between autophagy, cancer, and MAMs is not so difficult. Several findings may suggest this. For example, the corrected maintenance of MAMs integrity by MFN-1/-2 and DRP1 is also crucial for autophagosome formation and tumor cell growth. (D) The autophagic regulator AMBRA-1 has been found to interact with both the oncogene BCL-2 and the tumor suppressor BECLIN-1. Again, ER stress mediated by UPR a potent autophagic activator: at the same time ER stress is a critical signal capable to drive cell death. Also, the activity of the main proteins involved in Ca2+ release and reuptake at MAM levels, SERCA and IP3R3, is reported to be involved in apoptosis and tumorigenesis. Interestingly, the functioning of these channels is intimately regulated by several oncogenes (like AKT) and tumor suppressor (such as p53), which are also involved in regulation of autophagy. Abbreviations: grp75, glucose-regulated protein 75; IP3R3, inositol 1,4,5-trisphosphate receptor type 3; MFN-1/-2, mitofusin-1/-2; Hk2, Hexokinase 2; PACS, phosphofurin acidic cluster sorting protein; VDAC, voltage-dependent anion-selective channel; Ca2+, calcium; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1 alpha; PTEN, phosphatase and tensin homolog; p66Shc, 66 kDa proto-oncogene Src homologous-collagen homolog; PKCβ, protein kinase C beta; PP2a, protein phosphatase 2; AKT, protein kinase B; SERCA, sarco/endoplasmic reticulum Ca2+ ATPase; MTOR, mechanistic target of rapamycin; PML, promyelocytic leukemia; UPR, unfolded protein response; PINK1, PTEN-induced putative kinase 1; PERK, protein kinase-like ER kinase; DRP1, dynamin-related protein; Ero1a, ER oxidoreductase 1 alpha; IP3R3, inositol 1,4,5-trisphosphate receptor type 3; AMBRA-1, Beclin1-regulated autophagy; BCL-2, B-cell lymphoma 2; BECLIN-1, BCL-2-interacting protein;, AMPK, 5′ adenosine monophosphate-activated protein kinase).