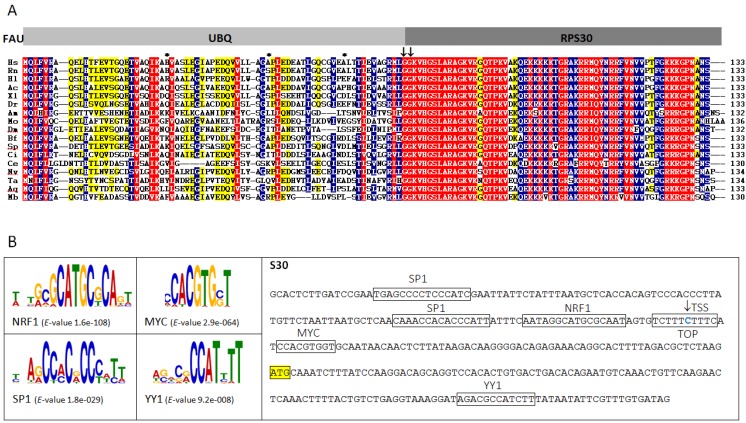
Figure 1.
(A) Multiple sequence alignment of FAU proteins from representative species (Homo sapiens (Hs), Rattus norvegicus (Rn), Haliaeetus leucocephalus (Hl), Anolis carolinensis (Ac), Xenopus laevis (Xl), Danio rerio (Dr), Apis mellifera (Am), Metaseiulus occidentalis (Mo), Drosophila melanogaster (Dm), Branchiostoma floridae (Bf), Strongylocentrotus purpuratus (Sp), Ciona intestinalis (Ci), Caenorhabditis elegans (Ce), Nematostella vectensis (Nv), Trichoplax adhaerens (Ta), Amphimedon queenslandica (Aq), Monosiga brevicollis (Mb)). G–G dipeptide motif and missing lysine residues which serve as sites for polyubiquitin chain formation are marked with arrows and asterisks, respectively; red = 100%, blue = 80% and yellow = 60% identity; (B) Over-represented motifs in sponge ribosomal protein gene (RPG) promoters and promoter sequence of sponge FAU with indicated translational start site (marked with yellow rectangle), transcription start site (TSS, marked with arrow) and transcription factor binding sites and terminal 5′-terminal oligopyrimidine (TOP) tract (marked with rectangles).