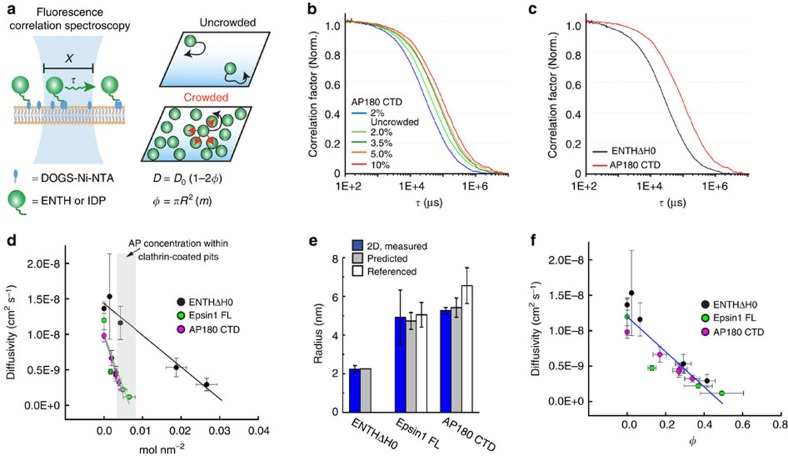
Figure 2. Intrinsically disordered proteins crowd membrane surfaces efficiently.
(a) Fluorescence correlation spectroscopy (FCS) is used to calculate molecular diffusivity by measuring the time (τ) that a fluorescent molecule (green spheres) takes to pass through a laser focal spot. Supported lipid bilayers (SLB) consisting of POPC lipids with increasing concentrations of DOGS-Ni-NTA lipids (blue ovals) were used to modulate the density of membrane-bound, histidine-tagged proteins. Diffusivity (D) decreases with molecular coverage, φ with the relationship D=Do(1–2φ). φ can be calculated as the projected area of a protein, πR2, multiplied by m, the number of molecules per nm2. (b) Normalized average FCS curves for AP180 CTD on SLB membranes of increasing molar % DOGS-Ni-NTA (2–10 mol%). (c) Normalized average FCS curves for ENTHΔH0 (black) compared with AP180 CTD (red). AP180 CTD has a slower time of diffusion despite similar number of molecules per area, ∼4 × 10−3 molecules per nm2. Experimental n values for all FCS data is described in methods. (d) Protein diffusivity as a function of the concentration of membrane-bound proteins. Error bars represent the mean±s.d. Experimental n values for all membrane coverage data is described in Supplementary Methods. (e) Calculated hydrodynamic radii, R, from 2D FCS and photon scanning measurements (b–d), predicted values (ENTH crystal structure or IDP polymer estimates) and referenced gel filtration/analytical centrifugation results11. Error bars are the mean±s.d. for 2D measurements, and for referenced measurements. Error bars for predicted sizes represent the range of persistence length values. (f) Diffusivity as a function of molecular occupancy, φ. All proteins collapse onto a single linear curve. Error bars represent the mean±s.d.