Figure 1. A very wide precursor ion (Open) search setting identified ~185,000 modified peptides.
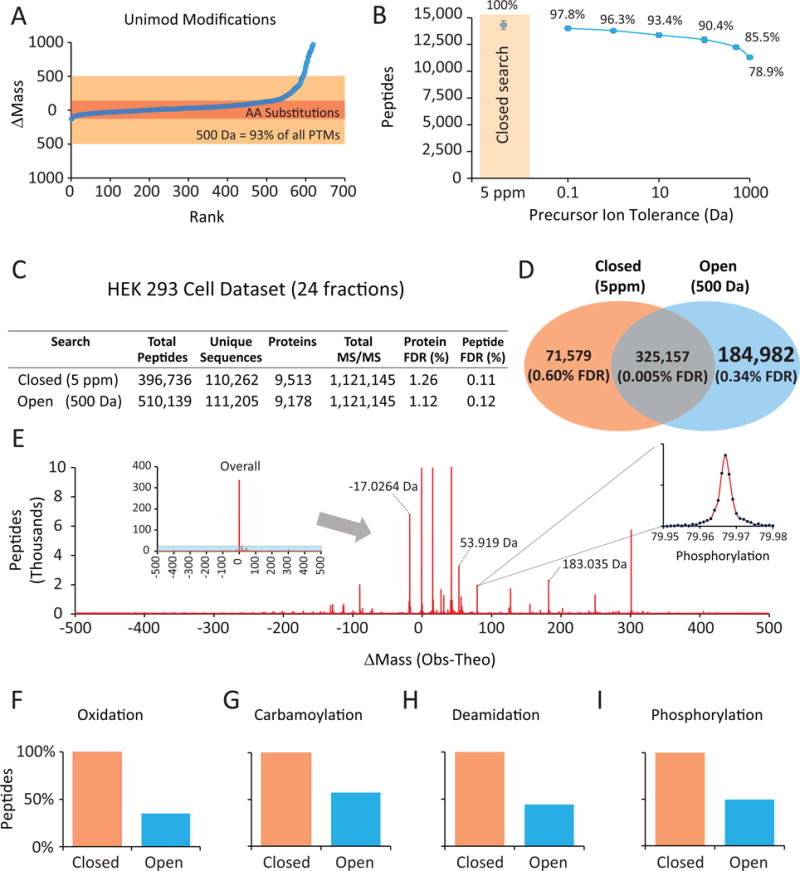
A) Most Unimod.org-reported modifications change precursor masses by < 500 Da. B) The vast majority of unmodified peptides are re-identified from Open searches. MS/MS spectra from mouse brain peptides analyzed in triplicate were searched using the Sequest algorithm and varying only the precursor ion tolerance. Note that fragment ion tolerance remained very strict (0.01 Da). At ±500 Da, 86% of unmodified peptides matched from an accurate mass search (5 ppm or ~.005 Da) were still assigned at a 1% FDR. C) A proteome-wide dataset was collected by LC-MS/MS from trypsinized and fractionated HEK293 cell lysate and assessed through either an Open or Closed search. D) The Open search identified more than 184,000 peptides with modified ΔM (mass change) values between −500 and +500 Da. E) ΔM distribution for 510,139 peptides. In addition to the 325,157 unmodified peptides, the 184,982 modified peptides distributed based on the exact net mass change of their modification. The inset shows a zoomed in view of ~2000 phosphorylated peptides. F–I) Comparison of identical modified peptides matched using a directed Closed search (where the modification was specified as differential) with an Open (±500-Da) search for four known peptide modifications: oxidation, carbamoylation, phosphorylation and deamidation.
