Figure 4. Analysis of ~185,000 peptides provided insights into rare biological modifications and amino acid variants/variations.
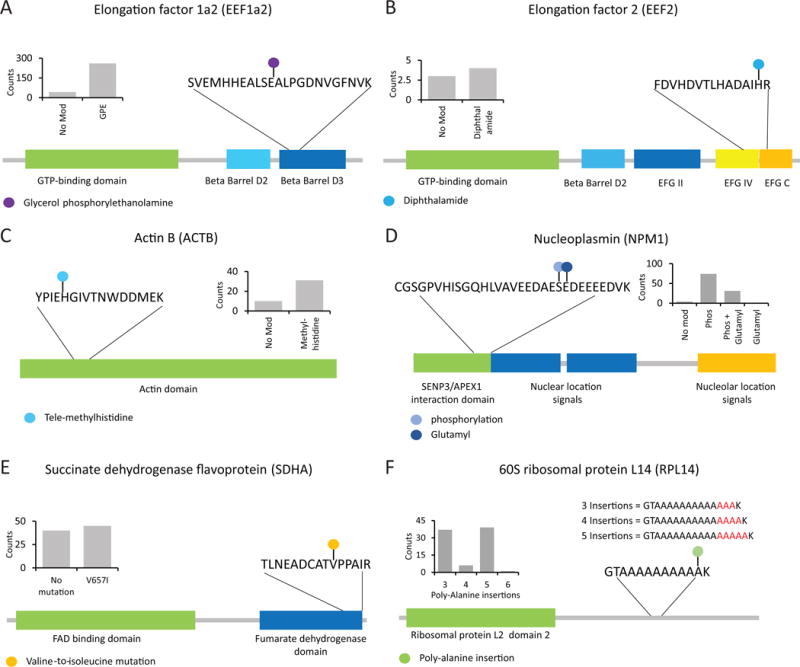
In each example, as shown in the inset, the modified form was detected more frequently than the matching unmodified peptide form. A) Glycerol phosphorylethanolamine modification of glutamate residue 301 in Elongation factor 1a2 was identified with hundreds of spectral counts. B) A diphthalamide modification was identified in position H715 for Elongation factor 2. C) A tele-methylhistidine modification was identified on cytoplasmic actin, ACTB. D) Nuceloplasmin was identified with two modifications, a phosphorylation event at S135 and glutamyl modifications at E136. The glutamylation event was only identified when phosphorylation was present giving a Δmass value of +209.0089 Da. E) One example from numerous amino acid variations identified in 293 cell Open searches. Succinate dehydrogenase with a V657I mutation is shown. F) Complex variations were also identified using the open search. For example, several alanine residues were added to the Ribosomal Protein L14 at position 159. These insertions numbered from as low as three to as many as six alanines.
