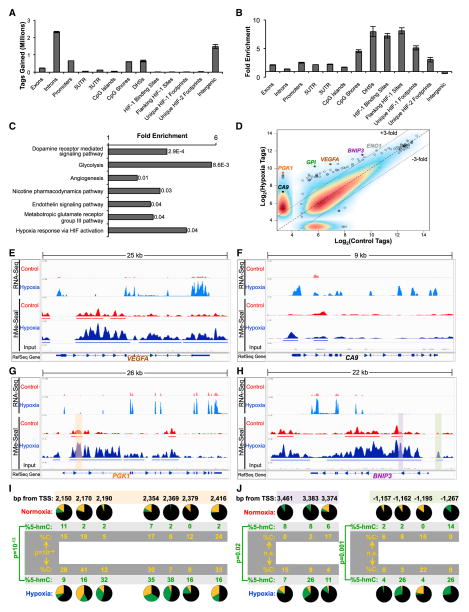
Figure 2. 5-hmC Gains Occur at Hypoxia Regulated Regions in SK-N-BE(2) Cells.
(A and B) Absolute 5-hmC gains (A) and enrichment for 5-hmC gains (B) were calculated at genomic elements. Regions flanking HIF binding sites are regions ±1 kb from the ChIP-seq peak. Footprints include the ChIP-seq peak and flanking region (Schödel et al., 2011). Data represent means ± SD for two biological replicates.
(C) PANTHER pathway analysis of genes with regions gaining at least 3-fold 5-hmC. p values are Bonferroni corrected results of binomial tests (Mi et al., 2013).
(D) Regions gaining the most 5-hmC map to the HIF-1 target genes CA9, PGK1, GPI, VEGFA, BNIP3, and ENO1. Plotted points represent combined data from two experiments.
(E–H) RNA-seq and hMe-Seal sequencing data are plotted in the Integrated Genomics Viewer (IGV) genome browser at VEGFA, CA9, PGK1, and BNIP3 genes. RefSeq gene tracks represent the position of known genes in the hg19 build of the human genome. Arrows represent the direction of transcription.
(I and J) TAB-seq data for the corresponding shaded regions of PGK1 (G) and BNIP3 (H). For each CpG, black, green, and gold represent the percentage of 5-mC, 5-hmC, and unmodified cytosine, respectively. Results represent pooled data over three independent experiments. p values calculated by chi-square tests. See also Figure S2 and Table S2.