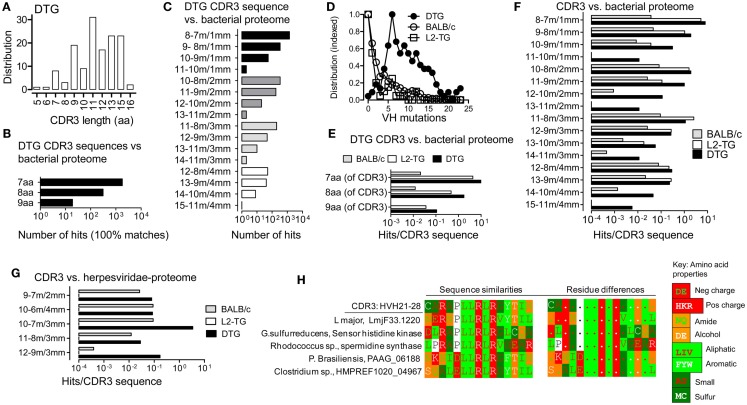
Figure 1.
Analysis of similarities between CDR3 sequences and the microbial proteome. (A) IgVH CDR3 length distribution of sequences from DTG mice with end stage lupus. (B,C,E–H), Results from Blastp analysis of 15mer CDR3 peptides vs. microbial proteomes and G, Herpesviridae proteome. (B) DTG IgG IgVH CDR3 peptides vs. microbial proteome: Numbers of hits with 100% matches with contiguous microbial sequences are shown (i.e. alignments without gaps). (C) DTG IgG IgVH CDR3 vs. microbial proteome: Number of hits in the indicated categories of length with matched/mismatched aa. For example: a 12 aa stretch that includes 10 matches (m) and 2 mismatches (mm) is labeled “12–10 m/2 mm.” (D) Mutations per IgVH sequence (not including CDR3) in the three sequence data sets: BALB/c, DTG IgG, L2-TG IgG (see Materials and Methods). The highest frequency of mutations for each data set is normalized to 1 on the Y axis. (E) Exact matches normalized to the number of input sequences in DTG vs. L2-TG IgG data sets. (F) Hits with matches/mismatches [as denoted in (C)] normalized to the number of input sequences in DTG, L2-TG, and BALB/c data sets. (G) Hits with matches/mismatches as in (F), compared to Herpesviridae proteome, normalized to the number of input sequences. (H) Example of one DTG anti-dsDNA IgVH CDR3 sequence and similar microbial sequences. An additional hit from outside the microbial databases is also shown, from the Leishmania major protozoa. Sequences similarities between IgVH CDR3 and proteins are shown in the left panels. Amino acids are color coded according to charge (Negative: D, E; positive: H, K, R), or the chemical properties of side chains (i.e. amide: N, Q; alcohol: S, T; aliphatic: L, I, V; aromatic: F, Y, W; small size: A, G; sulfur atom: M, C; or other: P), see key for color code. In the right panels, differences in the sequences are marked by aa symbols when such aa do not belong to the same chemical group as the most frequent aa at the corresponding position of the comparison set.