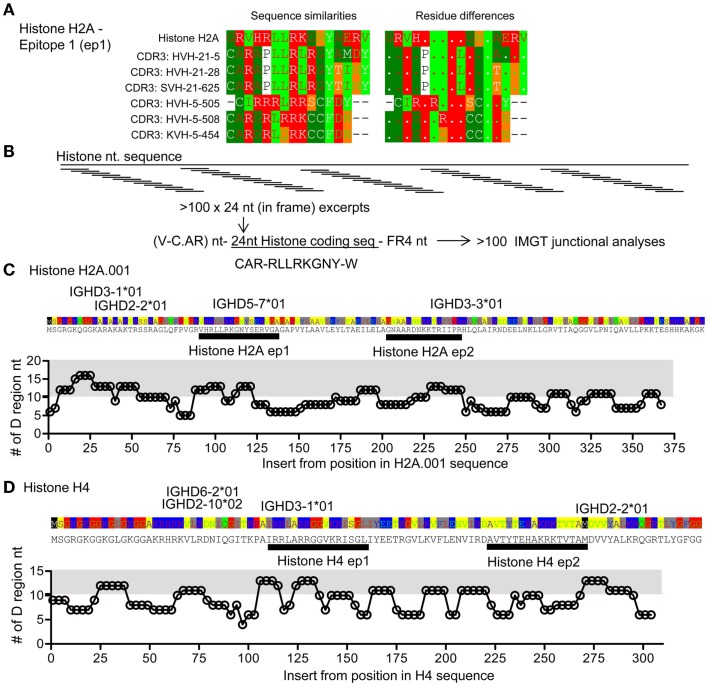
Figure 4.
Comparison of CDR3 and histone sequences; analysis of histone H2A sequence segments as part of V–D–J junctions. (A) Examples of sequence comparisons of IgVH CDR3 sequences from DTG mice with end stage lupus and histone H2A, this epitope is denoted epitope 1 H2A-ep1. See also Figure S1 in Supplementary Material. (B) Artificial hybrid sequence files were made by inserting histone sequences – replacing the IgVH CDR3 to form model sequences. IgVH CDR3 regions are usually less than eight amino acids long, corresponding to 24 coding nucleotides. In frame H2A sequences were electronically generated i.e. HisH2A nt 1–24, nt 4–27, … , nt 367–391 and electronically engrafted onto a random IgVH-segment. A random FR4 was grafted onto this artificial IgVH – “Histone H2A-CDR3.” Resulting model sequence files were up-loaded for IgVH junctional analysis at IMGT. (C) The lengths of suggested IMGT D-segments are plotted. The average D segment usage in BALB/c mice is 10.2 nt. Gray area: >11 nt. HisH2A sequence is provided above the plot, possible D gene segments are positioned according to highest levels of matching. Two epitopes are marked, histone H2A-ep 1 [see also (A)] and histone H2A-ep2. See also Figure S1 in Supplementary Material. (D) Analysis of histone H4 corresponding to that performed in (C). Two epitopes are marked histone H4-ep1 and histone H4-ep2, see also Figure 5 and Figure S1 in Supplementary Material.