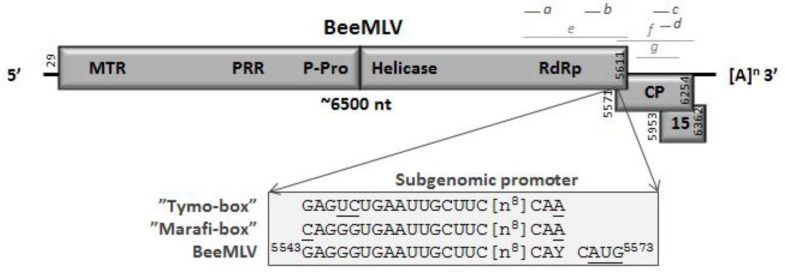
Figure 1.
Genome organization of BeeMLV. The estimated genome size is indicated as are the presence of a natural 3′ poly-A tail and the location of overlapping open reading frames coding for the capsid protein (CP), an unknown 15 kD protein (P15) and the large polyprotein containing the domains MTR (methyl transferase), PRR (Proline-rich region), P-Pro (endo-peptidase), Helic (NTPase/helicase) and RdRp (RNA-dependent RNA polymerase), separated by the location of the putative P-Pro cleavage site in between the P-Pro and Helic domains. Additionally indicated are the location and sequence of the putative BeeMLV sub-genomic RNA promoter, compared to its homologues for the Tymo- and Marifiviruses. The lines above the genome map identify the locations of the nucleotide (a) and RdRp (b) plus CP (c) amino acid sequences used in the phylogenetic analyses (light grey) and of the quantitative RT-qPCR assays for either genomic (d, e) or sub-genomic + genomic (f, g) virus RNA quantification (dark grey).