Fig. S3.
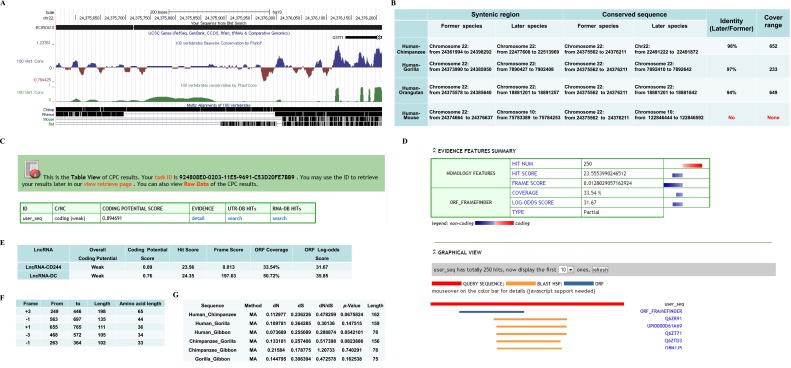
Bioinformatics analyses of evolutional conservation and protein-coding potential of lncRNA-BC050410. (A) The conservation track of lncRNA-BC050410 analyzed by the UCSC Genome Browser (genome.ucsc.edu). No mouse or rat homolog was found. (B) Conservation analysis in the syntenic region. Identity was calculated by alignment between later (e.g., chimpanzee) and former (e.g., human) species in the cover range (e.g., 652) indicated in the table. Note that no identity between human and mouse was found. (C and D) The analysis of protein-coding potential of lncRNA-BC050410 using tools provided by the Peking University Center for Bioinformatics (cpc.cbi.pku.edu.cn/programs/run_cpc.jsp) shows lack of protein-coding capability. (E) Comparison between lncRNA-BC050410 and the variant-1 transcript of lncRNA-DC (35) for parameters to determine protein-coding potential using the same tools provided by the Peking University Center for Bioinformatics (cpc.cbi.pku.edu.cn/programs/run_cpc.jsp). The larger hit score and frame score indicate that a transcript is more likely protein-coding, and larger ORF coverage or log-odds score indicates the better ORF quality. (F) The lengths of five potential ORFs in lncRNA-BC050410. (G) dN/dS analyses between two different species of Hominoidea indicated in the table. Data were calculated by the KaKs_Calculator software (https://code.google.com/p/kaks-calculator/wiki/KaKs_Calculator) using pair-wised DNA sequences (lengths are indicated in the table) from homologous amino acid sequences aligned between two different species of Hominoidea. “MA” indicates model averaging, which assigns each candidate model a weight value and engages more than one model to estimate average parameters across models (64). A value of P > 0.05 was considered as no negative or positive selection.
