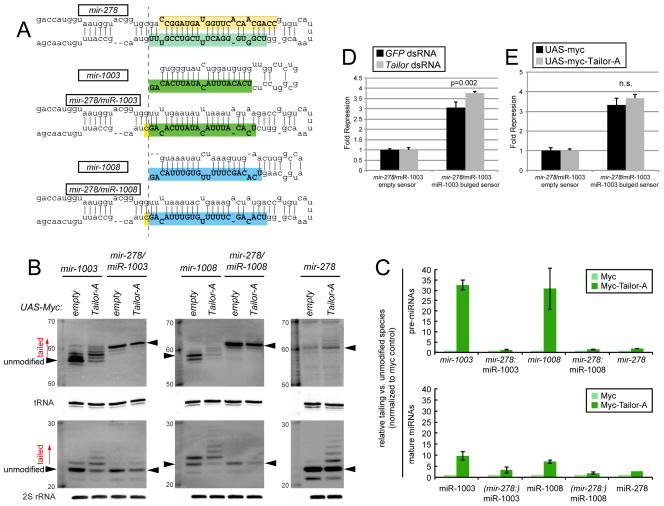
Figure 4.
Impact of biogenesis strategy on substrate recognition by Tailor. (A) The mature-3p species of mirtrons mir-1003 and mir-1008 were reprogrammed into the canonical mir-278 backbone. (B) Northern analysis of natural and reprogrammed mirtron constructs transfected into S2 cells, with or without Tailor expression construct. Robust tailing of mirtron hairpins and mature products is abrogated when their sequences are directed through the Drosha pathway. (C) Quantification of replicate experiments; SD is shown. The ratios of tailed to unmodified species (marked with arrowheads in B) in the presence of ectopic Tailor were normalized to parallel myc control conditions. The analysis shows mirtrons are preferred Tailor substrates, and their hairpins accumulate more tailed species than their mature miRNAs. (D, E) Reporter assays in S2 cells transiently expressing ub-Gal4, UAS-miRNA and the indicated luciferase sensors. (D) Activity of the reprogrammed mir-278/miR-1003 construct is less enhanced in Tailor-depleted cells than wildtype mir-1003 (Figure 3F). (E) Reprogrammed mir-278/miR-1003 construct is not inhibited by Tailor overexpression, as is wildtype mir-1003 (Figure 3G). Error bars represent SD and unpaired T-tests were used to calculate significance.