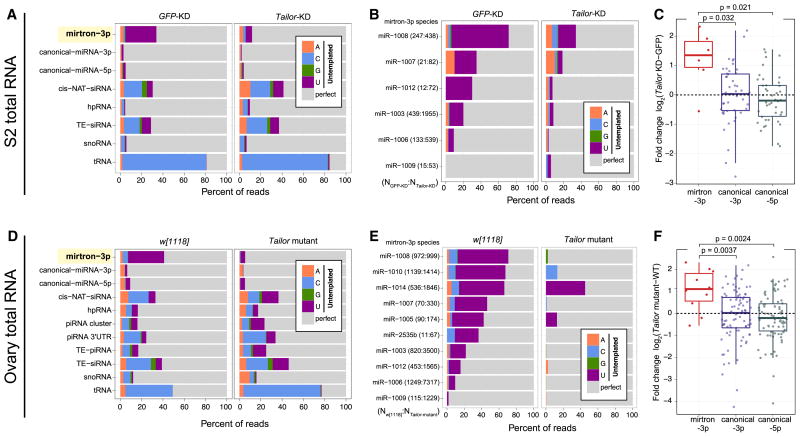
Figure 5.
Genomewide analysis of Tailor substrates in cultured cells and ovaries. Genome-matching reads are represented in gray while different untemplated nucleotides are color-coded. (A) Comparison of control and Tailor-depleted S2R+ cells demonstrates that mirtron-3p species are heavily uridylated, that this is nearly completely dependent on Tailor, and that other substrates are little affected by Tailor. (B) Analysis of individual mirtron-3p species shows their uridylation depends on Tailor. The number of mirtron reads in each library is shown in parentheses. (C) Expression levels of miRNAs in the presence and absence of Tailor. Most mirtron-3p species increase in Tailor-knockdown cells, whereas canonical miRNAs are not directionally affected. (D) Comparisons of w[1118] and Tailor-KO ovaries similarly shows that mirtron-3p species are the dominant class of Tailor-dependent, uridylated substrate. (E) Analysis of individual mirtron-3p species shows their Tailor-dependent modification. (F) Most mirtron-3p species increase in Tailor-knockout ovaries, whereas canonical miRNA species are not directionally affected. For analyses in A and D, all reads mapped to each sRNA class were pooled. For analyses in B–C and E–F, we used only mirtrons with ≥10 3p reads in both datasets under comparison. The box plots in C and F follow Tukey’s standard convention; significance was measured by t-test. See also Figure S4.