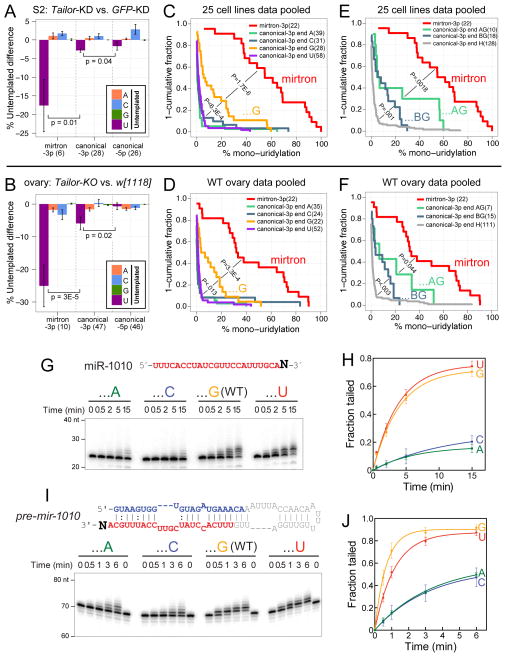
Figure 6.
Intrinsic activity of Tailor for 3′-G hairpin substrates. (A, B) Analysis of miRNAs with ≥10 reads and ≥1% untemplated uridylation in both paired datasets under comparison. In addition to the dominant uridylation of mirtron-3p reads, canonical miRNA-3p reads exhibit a greater degree of Tailor-dependent untemplated uridylation than do canonical miRNA-5p reads, in S2R+ cells (A) and ovaries (B). Wilcoxon rank-sum test (one-sided) was used to measure statistical significance; mean±SEM shown. (C–F) Analysis of miRNAs with ≥10 reads in the pooled cell line or ovary datasets (see Table S1). Wilcoxon rank-sum one-sided tests, incorporating multiple testing correction by Holm method, measured statistical significance of uridylation fraction enrichment between indicated small RNA classes. (C, D) Untemplated uridylation of mirtrons and canonical miRNA-3p species segregated by terminal nucleotide. While mirtrons exhibit far greater uridylation than canonical miRNAs, those canonical miRNA-3p species ending in G acquire greater uridylation than other miRNA subsets. (E, F) Terminal dinucleotide analyses of canonical miRNA-3p species and their uridylation. miRNAs ending in 3′-AG are more uridylated than other terminal G miRNAs (GG, CG, UG, i.e. “BG”), which in turn acquire greater uridylation than other terminal identities (A, U, C, i.e. “H”). This is true in both cell line (E) and ovary (F) data. (G–J) In vitro tailing assays of substrates bearing different 3′ nucleotides. To highlight substrate preferences, these assays utilize immunopurified Tailor-B material from stable cells, which is not as active as those in Figure 3. (G, H) Tailing assays on mature miR-1010 3′ variants. Tailor is most active on the normal 3′-G isoform, and also exhibits strong activity on the 3′-U variant. (I, J) Tailing assays on pre-mir-1010 3′ variants. Tailor maintains 3′-nt selectivity on hairpins, and tails them more effectively than mature miRNAs. See also Figure S5 and Tables S2 and S3.