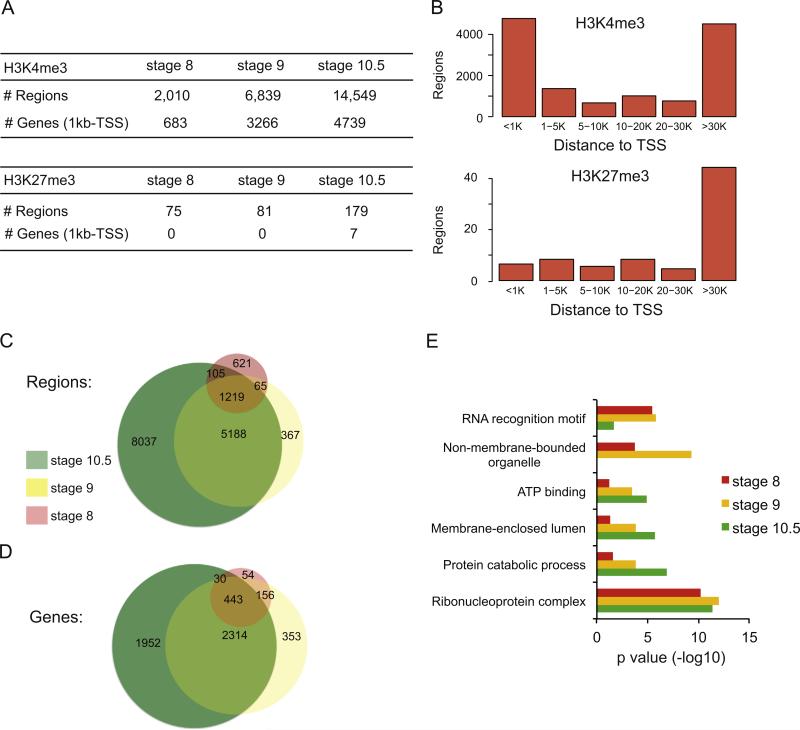
Fig. 1.
Occupancy of H3K4me3 and H3K27me3 in X. tropicalis at stage 8, 9 and 10.5. (A) Table showing the breakdown of numbers from the ChIP-Seq datasets for H3K4me3 (top) and H3K27me3 (bottom), including the number of regions identified and the genes that could be associated to the regions. Each category is depicted for stages 8, 9 and 10.5. (B) Histograms showing where the regions bound by either H3K4me3 (top) or H3K27me3 (bottom) exist with respect to annotated TSS regions at stage 10.5. The number of bound regions is plotted on the Y axis, with the distance from nearest TSS along the X axis. (C) Venn diagram showing how the regions bound to H3K4me3 compare between stage 8, 9 and 10.5. (D) Venn diagram showing how the genes associated with H3K4me3 compare between stage 8, 9 and 10.5. (E) DAVID analysis for genes associated with H3K4me3 at stage 8, 9 and 10.5 (red, yellow and green, respectively).