Figure.
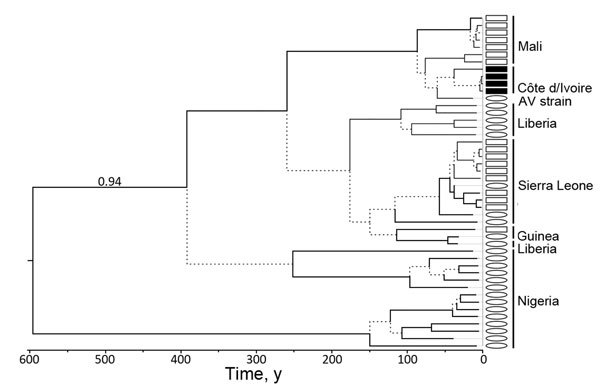
Bayesian chronogram of Lassa virus (LASV) sequences determined on the basis of a fragment of the large genomic segment. Branches receiving posterior probability values <0.95 and bootstrap values <50 (poorly supported) are dashed. LASV sequences of human origin are indicated by ovals, and those of multimammate rats are indicated by squares. Sequences reported in this study are indicated by black squares. This tree was built under the assumption of a molecular clock and is therefore rooted. The numerical value on the tree’s most basal branch is the root posterior probability of this branch; it supports the notion that LASV sequences from Nigeria and other countries are not reciprocally monophyletic. GenBank accession nos. of sequences used for phylogenetic analyses are shown in Technical Appendix Table 2. AV strain indicates the strain from a German patient.
