Figure 2. miRNA Profiling Workflow.
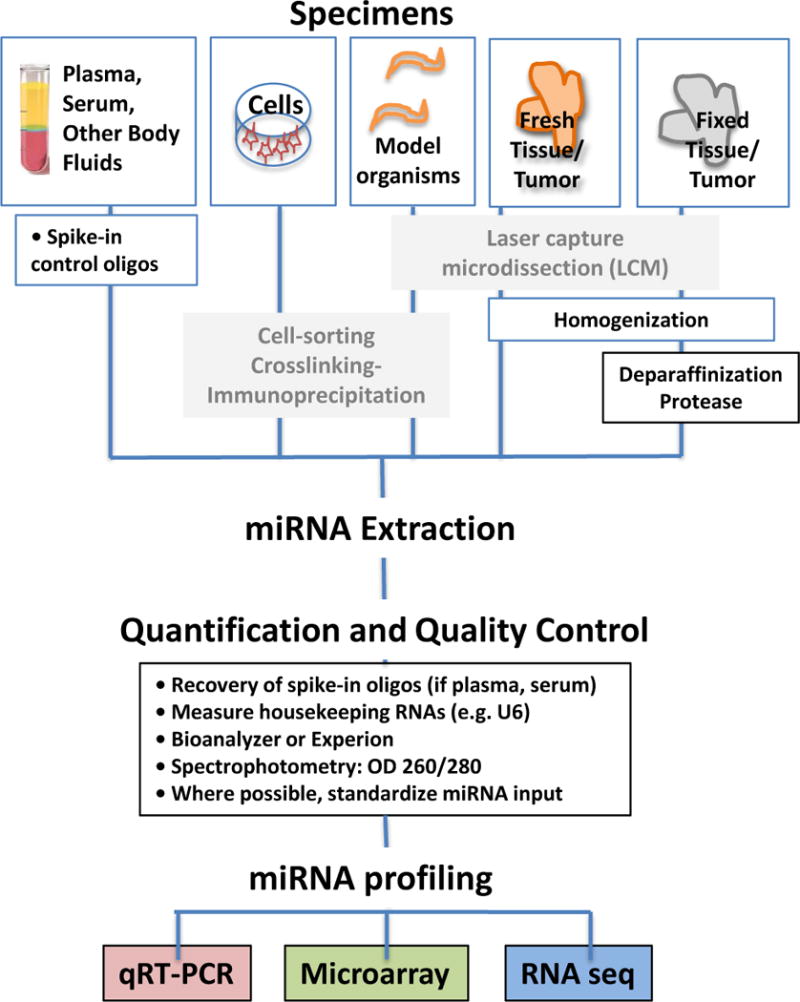
A) MicroRNAs can be extracted from a variety of specimen types, highlighted here are plasma and serum, cells in culture, fresh tissue/tumor, or fixed tissue/tumors. B) Methods for purifying miRNA populations of interest include size purification by gel electrophoresis, and AGO2 immunoprecipitation (AGO2-IP), with or without UV crosslinking (CLIP). Laser capture microdissection (LCM) can be used to purify material from fresh-frozen or formalin-fixed paraffin embedded (FFPE) tissue sections. C) MicroRNA isolation methods are similar to total RNA isolation, and commercially available extraction kits are listed in the light orange boxes. These typically use a chemical extraction combined with a purification step involving binding and eluting from a silica column. Formalin fixed (FF), or formalin fixed, paraffin embedded (FFPE) tissue/tumor require additional deparaffinization and protease treatment, and commercially available kits designed for these sample types are also listed in the light orange boxes. We recommend adding spike-in oligos prior to RNA extraction to enable a quality control check, particularly when using plasma or serum samples. D) A variety of methods can be used to assess miRNA quality after extraction. For most samples, these methods include spectrophotometry, automated capillary electrophoresis with the Bioanalyzer or Experion, and/or determining expression of housekeeping miRNAs. For serum and plasma, which usually have total RNA yields too low to accurately quantify, determining the recovery of spiked-in oligos can be a useful surrogate.
