Fig. 3. Representative AR genes captured through functional metagenomic selection of Amerindian oral and fecal microbiota.
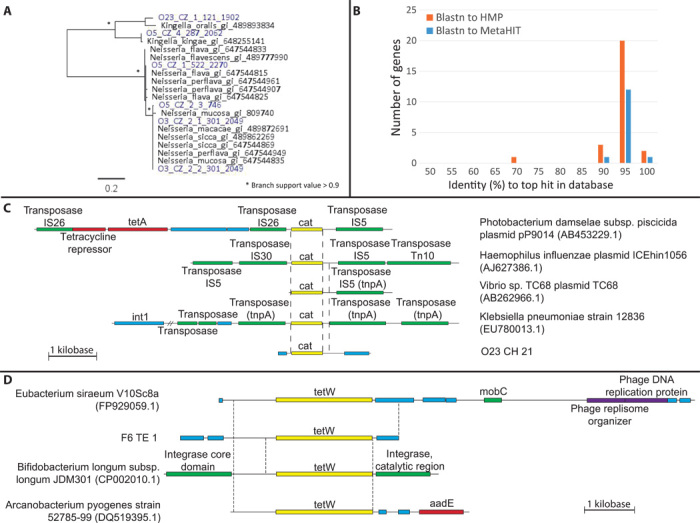
(A) Maximum likelihood (ML) tree of ceftazidime-resistant PBPs from Amerindian oral microbiota (blue) and their top blastx hits in NCBI nr (black). (B) Identity (%) of the 28 AR genes to their top hits in HMP and MetaHIT (blastn). Fourteen aligned to MetaHIT, all isolated from fecal microbiota. (C) Chloramphenicol acetyltransferase (cat) from O23_CH_21 and homologs in NCBI nt (nucleotide database). Dashed lines indicate 99% identical sequences. Dashes indicate omitted sequence (8487..25716). (D) tetW from F6_TE_1 and homologs in NCBI nt. Dashed lines indicate 98 to 99% identical sequences. Sequences are to scale. Yellow, AR gene; red, other resistance genes; green, mobile genetic elements; purple, bacteriophage genes; blue, other.
