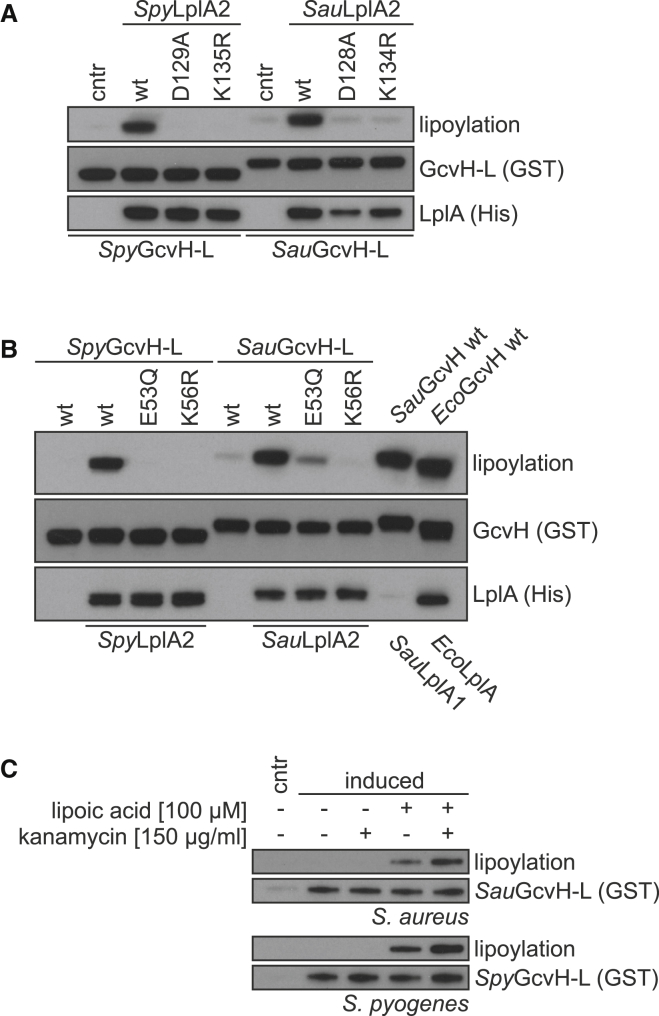
Figure 2.
The LplA2/GcvH-L Pair Resembles Its Canonical Homologs
(A) Wild-type, but not mutant LplA2, can lipoylate the GcvH-L protein. The mutations were chosen in analogy to the canonical LplA/GcvH pair of E. coli where they interfere both with the lipoate adenylation and the subsequent lipoate transfer (Fujiwara et al., 2010).
(B) Mutations of lipoylation motif residues within GcvH-L impair the lipoate transfer reaction. Mutation of Lys56 interferes with lipoyl attachment, whereas Glu53 is important for recognition by LplA2 (Fujiwara et al., 1991, 2010). Control reactions were carried out using the canonical LplA/GcvH pairs of S. aureus (SauGcvH, SauLplA1) and E. coli (EcoGcvH, EcoLplA).
(C) SauGcvH-L and SpyGcvH-L were expressed in the presence and absence of lipoic acid supplementation. In addition, protein synthesis of some samples was interrupted by supplementation with kanamycin 1 hr prior to culture harvesting. The effect of the additives on GcvH-L lipoylation was assessed by immunoblot. For further characterization of the LplA2/GcvH-L pair, see Figure S2.