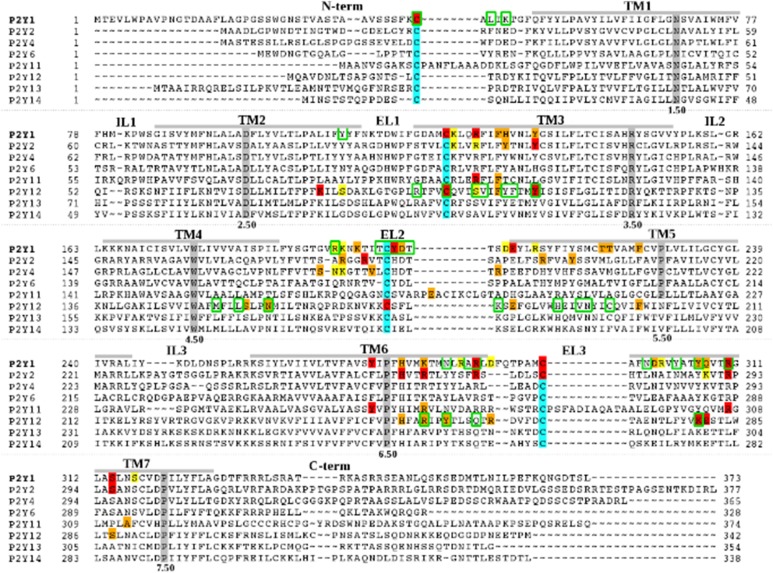
Fig. 2.
Sequence alignment of the human P2YRs. Residues that have been identified using site-directed mutagenesis, as involved in ligand binding and/or receptor activation at P2Y1R (Abbracchio et al., 2006; Zhang et al., 2015), P2Y2R (Erb et al., 1995; Hillmann et al., 2009), P2Y4R (Herold et al., 2004), P2Y11R (Zylberg et al., 2007), and P2Y12R (Hoffmann et al., 2008; Mao et al., 2010; Ignatovica et al., 2012; Zhang et al., 2014a,b) are highlighted with different colors: residues whose mutation can have a major effect on ligand binding and/or receptor activation (red); residues whose mutation modulates ligand binding and/or receptor activation (orange); and residues whose mutation has a minor or no effect on ligand binding and/or receptor activation (yellow). Residues within 3 Å from the crystallographic pose of 2MeSADP at P2Y12R or within 3 Å from the crystallographic pose of MRS2500 at P2Y1R are circled in green. The most highly conserved residue among GPCRs of each helix is highlighted in gray. Cysteine residues involved in disulfide bridges are highlighted in cyan.