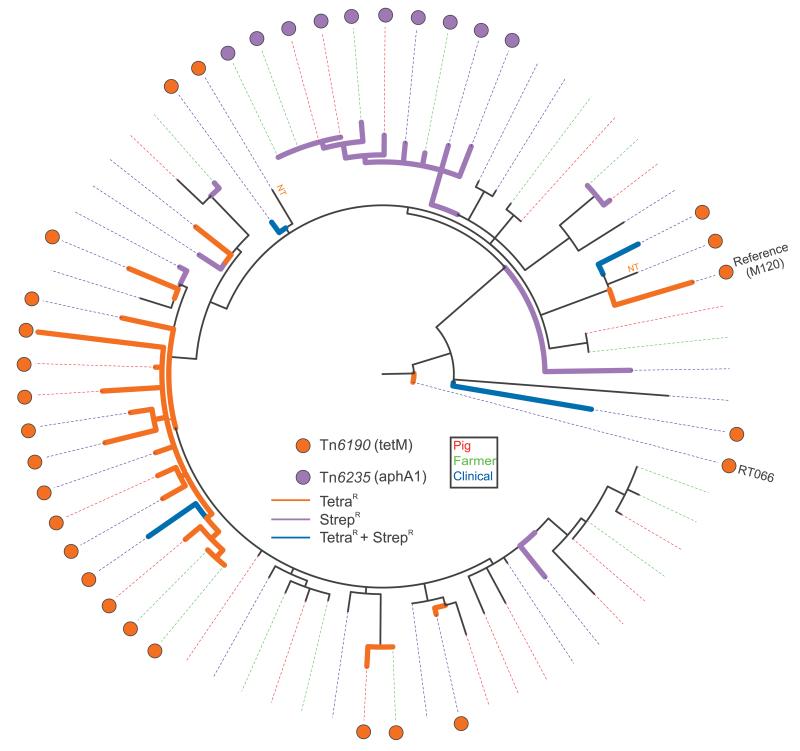
Figure 3. Phylogeny of Clostridium difficle 078 isolates showing the presence of antimicrobial resistance determinants, the Netherlands, 2002–11 (n=65a).
NT = not phenotypically tested.
Circular representation of the C. difficile 078 phylogeny with coloured dots representing the distribution of antimicrobial resistance (AMR) determinants. The legend shows the identified transposons together with the AMR determinants (between brackets) located on the transposon. The coloured dotted lines represent the source of the respective isolates (swine, farmer and clinical isolate). The presence of Tn6190 (tetM) is associated with tetracycline resistance; 078 isolates phenotypically tested as tetracycline-resistant are indicated with orange tree branches, streptomycin-resistant isolates are indicated with purple tree branches, isolates resistant to both tetracycline and streptomycin are indicated with blue tree branches.
a Two isolates were sequenced in duplicate. Ne RT 066 sequence was included as root sequence. In total, 68 sequences are shown.