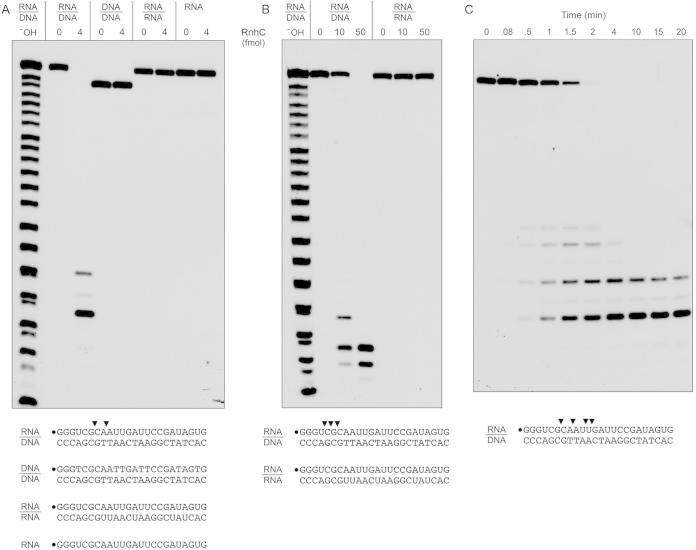
FIG 3.
Substrate specificity and kinetics. (A and B) Reaction mixtures (10 μl) containing 25 mM Tris-HCl (pH 7.5); 50 mM NaCl; either 10 mM MgCl2 (A) or 10 mM MnCl2 (B); 1 mM DTT; either 200 fmol 32P-RNA:DNA, 32P-DNA:DNA, or 32P-RNA:RNA duplexes or 200 fmol 32P-RNA single strand (as depicted at the bottom, with the 5′ radiolabel denoted by a dot); and RnhC as specified were incubated at 37°C for 20 min. The products were analyzed by urea-PAGE and visualized by autoradiography. An alkaline hydrolysis ladder of the 32P-labeled 24-mer RNA strand was analyzed in parallel in lanes −OH. (C) Time course of RNA:DNA cleavage. Reaction mixtures (110 μl) containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 20 nM 32P-RNA:DNA hybrid duplex (depicted at the bottom), and 0.4 nM RnhC were incubated at 37°C. Aliquots (10 μl) were withdrawn at the times specified and quenched with formamide-EDTA. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. The principal sites of RNase H incision are indicated by arrowheads above the 32P-labeled RNA strands shown at the bottom.