ABSTRACT
In bacteria, copper homeostasis is closely monitored to ensure proper cellular functions while avoiding cell damage. Most Gram-positive bacteria utilize the copYABZ operon for copper homeostasis, where copA and copB encode copper-transporting P-type ATPases, whereas copY and copZ regulate the expression of the cop operon. Streptococcus mutans is a biofilm-forming oral pathogen that harbors a putative copper-transporting copYAZ operon. Here, we characterized the role of copYAZ operon in the physiology of S. mutans and delineated the mechanisms of copper-induced toxicity in this bacterium. We observed that copper induced toxicity in S. mutans cells by generating oxidative stress and disrupting their membrane potential. Deletion of the copYAZ operon in S. mutans strain UA159 resulted in reduced cell viability under copper, acid, and oxidative stress relative to the viability of the wild type under these conditions. Furthermore, the ability of S. mutans to form biofilms and develop genetic competence was impaired under copper stress. Briefly, copper stress significantly reduced cell adherence and total biofilm biomass, concomitantly repressing the transcription of the gtfB, gtfC, gtfD, gbpB, and gbpC genes, whose products have roles in maintaining the structural and/or functional integrity of the S. mutans biofilm. Furthermore, supplementation with copper or loss of copYAZ resulted in significant reductions in transformability and in the transcription of competence-associated genes. Copper transport assays revealed that the ΔcopYAZ strain accrued significantly large amounts of intracellular copper compared with the amount of copper accumulation in the wild-type strain, thereby demonstrating a role for CopYAZ in the copper efflux of S. mutans. The complementation of the CopYAZ system restored copper expulsion, membrane potential, and stress tolerance in the copYAZ-null mutant. Taking these results collectively, we have established the function of the S. mutans CopYAZ system in copper export and have further expanded knowledge on the importance of copper homeostasis and the CopYAZ system in modulating streptococcal physiology, including stress tolerance, membrane potential, genetic competence, and biofilm formation.
IMPORTANCE S. mutans is best known for its role in the initiation and progression of human dental caries, one of the most common chronic diseases worldwide. S. mutans is also implicated in bacterial endocarditis, a life-threatening inflammation of the heart valve. The core virulence factors of S. mutans include its ability to produce and sustain acidic conditions and to form a polysaccharide-encased biofilm that provides protection against environmental insults. Here, we demonstrate that the addition of copper and/or deletion of copYAZ (the copper homeostasis system) have serious implications in modulating biofilm formation, stress tolerance, and genetic transformation in S. mutans. Manipulating the pathways affected by copper and the copYAZ system may help to develop potential therapeutics to prevent S. mutans infection in and beyond the oral cavity.
INTRODUCTION
Many bacteria utilize copper as an essential cofactor for enzymes involved in electron transfer reactions, including superoxide dismutase, cytochrome c oxidase, and NADH dehydrogenase (1, 2). Although it is required in low concentrations, copper at higher levels can pose a threat because of its high reactivity. Copper can exist in cuprous (Cu+) and cupric (Cu2+) oxidative forms; the interchange between these states via Fenton reactions can lead to the generation of reactive oxygen species (ROS), especially superoxide radicals. The ROS are believed to cause cellular damage by reacting with a number of cellular macromolecules, such as lipids, proteins, and nucleic acids (3–5). However, there is little or no direct evidence that shows that copper-induced ROS generation is the main mode of copper toxicity in bacteria (6–8). In Escherichia coli, copper was demonstrated to target and replace iron from the iron-sulfur clusters of dehydratases, thereby rendering them inactive and perturbing the biosynthesis of branched-chain amino acids (6–9). Supplementation of E. coli cultures with branched-chain amino acids was shown to reverse the copper-induced growth inhibition (7). However, a similar mechanism was not observed in Salmonella enterica serovar Typhimurium, where the addition of exogenous branched-chain amino acids did not revert copper-mediated growth inhibition, thereby suggesting the involvement of a different cellular target for copper toxicity (10). Copper-mediated membrane depolarization has also been reported in E. coli (11). The challenges posed by copper necessitate the involvement of complex regulatory machinery to maintain copper homeostasis in the cell. To contend with copper toxicity, bacteria utilize at least one of three principal mechanisms, including (i) copper export across the plasma membrane into the periplasmic space or the extracellular environment, (ii) extracellular and/or intracellular copper sequestration via copper binding proteins, and (iii) copper oxidation to a less toxic Cu2+ state (12). The mechanisms involved in copper stress tolerance can vary among bacterial species, and exploring these pathways is an important step to implement the use of such metal cations as antimicrobial therapeutics.
Streptococcus mutans is considered the primary etiological agent of dental caries, one of the most widespread infectious diseases worldwide (13, 14). Numerous studies have indicated the inhibitory effects of copper on S. mutans growth and caries formation (15–22). The copper concentrations in saliva and dental plaque vary between individuals and depend on the age, sex, nutrient intake, etc. (23–26). Copper concentrations in saliva can range from 0.05 to 61.7 μM, whereas the concentrations fluctuate between 26 and 1,520 ppm within the plaque biofilm. By the administration of copper-containing mouth rinses, copper has been shown to dramatically influence the growth and pathogenicity of S. mutans (25). In S. mutans strain GS-5, copper irreversibly inhibited the activity of F-ATPases, thereby compromising the cell's ability to carry out glycolysis in acidic environments (27). Furthermore, this cation was shown to modulate the expression of the glucosyltransferase D gene (gtfD), whose products are associated with soluble glucan production and biofilm formation in S. mutans (28). Like the genomes of many Gram-positive bacteria, the genome sequence of S. mutans does not appear to encode proteins or enzymes that require copper for their functional activity (29). Even though copper is not required for its cellular processes, S. mutans must still possess a functional copper-exporting machinery to avoid cell damage under excessive copper concentrations.
Unlike Gram-negative bacteria, which traffic copper through different cellular compartments from the cytosol to the periplasm and from the periplasm to the extracellular environment, Gram-positive bacteria possess rather simple copper homeostatic systems dedicated to extrusion of excess copper cations from the cytosol to the extracellular milieu (30). The CopYABZ copper homeostasis system of Enterococcus hirae is the best understood copper homeostasis model in Gram-positive bacteria (30–33). This operon encodes four proteins that include two copper P-type ATPases, CopA and CopB, a copper-responsive repressor, CopY, and a copper chaperone, CopZ (31–33). The CopZ family of metallochaperones is conserved within various bacterial and eukaryotic systems (30, 33). A homologous copper transport and resistance system, encoded by copYAZ, has been partially characterized in S. mutans strain JH1005 (34). The sensitivity of the copYAZ knockout mutant was shown to be specific to copper, and CopY was demonstrated to act as a negative regulator of the operon (34). Although the idea was not tested conclusively, it was speculated that CopZ derepressed the cop operon activity, and a deficiency of copYAZ was shown to result in enhanced sensitivity to cell killing under copper stress (34).
Here, we investigated the effects of copper on the physiology of S. mutans and the role of CopYAZ in copper homeostasis. While previous studies have implicated metal cations in biofilm formation and the development of genetic competence (35–37), the role of copper and the CopYAZ system in regulating these phenotypes in S. mutans is poorly understood. The ability of this organism to adhere to hard surfaces and form a biofilm is an important virulence factor that is critical for its survival and persistence in the oral cavity. Within the plaque biofilm, S. mutans is capable of natural transformation that is made possible when it accomplishes a transient physiological state referred to as genetic competence (38, 39). Genetic transformation is important for acquiring novel, heritable functions that can enhance fitness and drive evolution (40). In S. mutans, genetic competence is induced by two signaling peptides, designated the competence-stimulating peptide (CSP; encoded by comC) and the comX-inducing peptide (XIP; encoded by comS) (41–44). Under specific competence-inducing conditions, both CSP and XIP can activate the transcription of the master competence regulator that is encoded by the comX (or sigX) alternate sigma factor and is required for the transcription of genes involved in DNA uptake and recombination (41–44). Notably, the S. mutans competence development pathway activated by CSP and XIP is closely intertwined with its biofilm pathway (38, 41, 45–47). However, it remains to be studied how and whether copper exerts an influence on the regulation of these phenotypes via the putative CopYAZ copper transport system in S. mutans.
Here, we show that copper instigates a compromised state in S. mutans by dissipating membrane potential and decreasing the organism's ability to endure environmental oxidative and pH stress, produce biofilm, and develop genetic competence. Utilizing a copYAZ deletion mutant, we validated the function of S. mutans CopYAZ in copper efflux. We also report that the addition of copper or the absence of the copYAZ system reduces the transcription of genes involved in biofilm matrix production and the development of genetic competence; loss of copYAZ leads to impairment of stress tolerance, transformability, and membrane potential in S. mutans.
MATERIALS AND METHODS
Strains and growth conditions.
All S. mutans strains were grown in Todd-Hewitt yeast extract (THYE) (Becton Dickinson, Sparks, MD) broth as standing cultures or on THYE medium with 1.5% (wt/vol) agar (Bioshop; Burlington, Ontario, Canada) at 37°C in air with 5% (vol/vol) CO2. Tryptone yeast extract (TYE) medium (10% tryptone [Bioshop, Burlington, Ontario, Canada], 5% yeast extract, 17.2 mM K2HPO4) was utilized for the acid tolerance response (ATR) assays. For the ATR assays, NaOH or HCl was added to TYE medium to adjust the pH to 7.5 or 5.5 and 3.5, respectively. E. coli strains were cultivated aerobically in Luria-Bertani (LB) medium at 37°C. Chemically defined medium (CDM) (43, 48) was used for transformation frequency assays. Antibiotics were added whenever required at the recommended concentrations as follows: erythromycin (10 μg/ml), chloramphenicol (10 μg/ml), or spectinomycin (1,000 μg/ml) for S. mutans and ampicillin (100 μg/ml) or chloramphenicol (20 μg/ml) for E. coli.
MIC assays.
MIC assays were conducted as previously described (49). Briefly, 100 μl of mid-log-phase bacterial cells adjusted to an optical density at 600 nm (OD600) of ∼0.01 were added to a 96-well microtiter plate containing THYE medium supplemented with 2-fold serial dilutions of CuSO4 or CuCl2 (Sigma-Aldrich) solution (concentrations ranging from 0 to 25 mM) and AgNO3 (1 to 100 μM), CdSO4 (0 to 1.5 μM), HgNO3 (0 to 10 μM), ZnCl2 (0 to 25 mM), MnCl2 (0 to 100 mM), or CaCl2 (0 to 100 mM). After incubation at 37°C with 5% (vol/vol) CO2 for 24 h, bacterial growth was spectrophotometrically measured by using a microtiter plate reader at an absorbance of 600 nm. The MIC was determined as the lowest concentration that inhibited visible cell growth relative to the growth in the no-copper control.
Mutant and complemented-strain construction.
A mutant with deletion of the copYAZ operon (NCBI database gene annotations, SMU_424, SMU_426, and SMU_427) in the S. mutans UA159 wild-type background was constructed for this study. Briefly, a knockout mutant was constructed using the PCR-ligation mutagenesis strategy as described by Lau et al. (50) by deleting the operon and inserting an erythromycin resistance cassette at the locus in the UA159 background. The deletion of the copYAZ operon in the ΔcopYAZ strain was confirmed by PCR amplification, DNA sequencing, and quantitative real-time PCR (qRT-PCR) analyses. The complementation analysis was done using an E. coli copA-deficient mutant (strain DW3110 from the Keio collection). Complemented strains of E. coli copA were constructed using the Thermo Scientific CloneJET PCR cloning kit according to the manufacturer's instructions. Briefly, S. mutans copYAZ was PCR amplified using UA159 genomic DNA as the template and ligated into the pJET vector. The vector was first transferred into E. coli DH5α chemically competent cells (Subcloning Efficiency DH5α competent cells; Invitrogen). The clones were selected on LB agar plates supplemented with ampicillin (100 μg/ml) and confirmed using nucleotide sequencing. The resulting plasmid was then transferred into chemically competent cells of the E. coli DW3110 mutant to obtain the complemented strain DW3110compYAZ. Similarly, to generate a complemented strain in S. mutans, similar PCR products were utilized and ligated into plasmid pIB166 (51), which harbors a chloramphenicol resistance marker and a constitutive promoter upstream from a multiple cloning site. The cloned vector with the entire copYAZ operon was transferred into the ΔcopYAZ mutant, and the resulting strain was designated CompΔcopYAZ. Complementation with the empty vector was utilized as a control [DW3110(pJET) or ΔcopYAZ(pIB) strain] (data not shown). Bacterial strains utilized in this study are summarized in Table 1, and primers are in Table S1 in the supplemental material.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. mutans strains | ||
| UA159 | Wild type; Erms | J. Ferretti, University of Oklahoma |
| ΔcopYAZ mutant | In-frame copYAZ deletion mutant derived from UA159; Ermr | This study |
| CompΔcopYAZ | ΔcopYAZ strain transformed with pIB-YAZ; Cmr Ermr | This study |
| E. coli strains | ||
| W3110 | Wild type | Keio Collection |
| DW3110 | W3110 ΔcopA | Keio Collection |
| DW3110compYAZ | DW3110 transformed with pJET-copYAZ; Ampr Emr | This study |
| Plasmids | ||
| pIB166 | Shuttle plasmid containing the P23 lactococcal promoter; Cmr | 51 |
| pIB-YAZ | S. mutans copYAZ fragment cloned into pIB166; Cmr Emr | This study |
| pJET | Plasmid containing T7 promoter; Ampr | Thermo Scientific CloneJET PCR cloning kit |
| pJET-copYAZ | S. mutans copYAZ fragment cloned into pJET; Ampr Emr | This study |
| pDL277 | E. coli-Streptococcus shuttle vector; Spr | 109 |
Biofilm assays.
Biofilms were cultivated in 96-well polystyrene microtiter plates using one-quarter-THYE medium supplemented with 10 mM sucrose and various concentrations of CuSO4 or CuCl2 (0, 100 μM, 250 μM, 500 μM, 1 mM, 2 mM, or 5 mM). All wells were inoculated with overnight cell suspensions of S. mutans wild-type and mutant strains, and the plates were incubated for 24 h at 37°C with 5% (vol/vol) CO2. Following incubation, the broth was carefully removed and the biofilms were stained with 0.01% (wt/vol) safranin to determine the relative biomass, as previously described (52). For biofilm initial adhesion studies, cells (20 μl of mid-logarithmic cells) were incubated in 12-well polystyrene microtiter plates containing 2 ml one-quarter-THYE medium supplemented with 10 mM sucrose and various concentrations of CuSO4 (0, 500, or 1,000 μM). The plates were incubated for different time intervals, and the planktonic cells were removed. Attached cells were washed twice with 1× phosphate-buffered saline (PBS) before being scraped from the plates and resuspended in 200 μl of PBS. Cells were briefly sonicated and serially diluted in 10-fold decrements in 1× PBS, and 20-µl amounts of each dilution were spotted in triplicates on THYE agar plates. After 48 h of incubation, colonies were counted for the dilutions showing individual colonies (30 to 300 in number), and the percentages of cells that were attached/able to grow in the presence and absence of copper were calculated.
ATR assays.
Acid tolerance response (ATR) assays were conducted as previously described (53). Briefly, overnight cultures grown in THYE medium were diluted 1:20 using sterile prewarmed tryptone-yeast extract medium at pH 7.5 and supplemented with 1% (wt/vol) glucose (TYEG medium). Cultures were then grown to mid-logarithmic phase (OD600 of ∼0.4), divided into two equal aliquots, and pelleted via centrifugation. For nonadapted cells, one aliquot was resuspended in TYEG medium at a lethal pH of 3.5, and for adapted cells, the other aliquot was first resuspended in TYEG medium at pH 5.5 and incubated at 37°C with 5% CO2 for 2 h before being exposed to the lethal pH of 3.5. Following incubation at 37°C with 5% CO2, cell fractions were removed from cultures at time zero and after 2 h. Cells were gently sonicated, serially diluted in 10 mM potassium phosphate buffer (pH 7.2), and spotted in triplicate (20 μl of each dilution) onto THYE agar plates. After incubation for 48 h, CFU were counted and ATR was calculated as the percentage of CFU obtained at lethal pH after 2 h relative to the number of CFU present at time zero.
Growth kinetics analysis.
For growth kinetics assays, S. mutans strains were grown in THYE medium with various concentrations of CuSO4 or CuCl2, paraquat (0, 5 mM, 10 mM, 25 mM, or 50 mM), or hydrogen peroxide (H2O2) (0, 0.0015, 0.003, 0.0045, or 0.006% [vol/vol]). Overnight cultures of S. mutans strains were diluted 1:20 in fresh THYE medium and grown until mid-logarithmic phase (OD600 of ∼0.4). Amounts of 20 μl of cell inocula were added to 350-μl volumes of THYE medium containing various concentrations of test reagents, in quadruplicate. Cells were incubated at 37°C for 24 h; uninoculated wells and wells containing THYE medium alone, with or without reagents, were used as controls. No antibiotics were used in growth assays to avoid additional stress. Optical density readings were taken every 20 min using an automated growth reader workstation (Bioscreen C; Growth Curves USA) and were plotted over time to obtain growth curves.
Copper transport assays.
Overnight cultures grown in THYE medium were diluted 1:20 and suspended in fresh medium supplemented with or without 1 mM CuSO4 or CuCl2 (suspended in MilliQ water). Samples were incubated at 37°C with 5% CO2 until they reached mid-logarithmic phase (OD600 of 0.4 to 0.6). Following incubation, cells were harvested by centrifugation at 3,800 × g. To minimize the probability of measuring membrane-associated copper cations, cells were washed twice with 0.01 M NaClO4, an experimental electrolyte previously shown to remove residual metal from medium and bacterial surfaces (54). Cells were resuspended in ice-cold 1× PBS and filtered through a 0.22-μm-pore-size nitrocellulose filter. The filter was washed twice with 1 ml of ice-cold 1× Tris-EDTA buffer and once with 1 ml of ice-cold MilliQ water. The filter was digested with 4% HNO3, followed by inductively coupled plasma-atomic emission spectroscopic (ICP-AES) analysis using the ANALEST facilities (Analytical Laboratory for Environmental Science Research and Training) at the University of Toronto. A 100 mg/ml CuSO4 or CuCl2 (suspended in 4% HNO3 in MilliQ water) stock solution was used to prepare standards ranging from 3 μg/ml to 100 μg/ml for ICP-AES calibration. A standard curve was plotted and utilized to analyze the intracellular copper concentrations. No detectable amounts of copper were observed in THYE or LB medium alone. The measured intracellular copper concentrations were normalized per mg dry weight of cells.
Quantitative real-time PCR assays.
Total RNA was isolated from 18-h-old biofilms grown in the presence or absence of 500 μM CuSO4 in one-quarter-THYE medium supplemented with 10 mM sucrose. Due to its poor biomass production, higher numbers of wells (three times the number for the wild type) were inoculated to isolate RNA from 18-h-old biofilms of the ΔcopYAZ strain grown in the presence of copper. The planktonic cells were removed, and the biofilm cells were washed with 1× PBS. The biofilm cells were scraped from the polystyrene surface and suspended in ice-cold 1× PBS. The cells were sonicated and collected after centrifugation at 3,800 × g. For expression analysis of the genes associated with the competence regulon, cells were cultivated in CDM (43, 48) at 37°C with 5% CO2 until they reached mid-logarithmic growth phase. Cells were then treated with 100 μM CuSO4 for 30 min. For expression analysis of the genes associated with acid stress, mid-logarithmic cells cultivated in THYE medium were treated with 500 μM CuSO4 for 30 min. Total RNA was harvested from the cell pellets as previously described (55). After DNase treatment, RNA was subjected to reverse transcription using a first-strand cDNA synthesis kit (Fermentas). Quantitative real-time PCR analyses were conducted using the QuantiTect SYBR green PCR kit (Qiagen, Mississauga, Ontario, Canada). The fold change in expression was calculated according to the method of Pfaffl et al. (56, 57) using the following formula: fold change = [Etarget gene (control CT − experimental gene CT)]/[E16S rRNA (control CT − experimental gene CT)], where E (10−1/slope) represents the efficiency of gene amplification and CT is threshold cycle (49, 55). The results were normalized against S. mutans 16S rRNA or gyrA expression; the expression of these housekeeping genes was found to be stable under the test conditions (data not shown). Statistical analysis was conducted using Student's t test, and a P value of <0.05 was considered significant. The primers utilized are listed in Table S1 in the supplemental material.
Membrane potential assays.
Overnight S. mutans cultures were diluted 1:20 in fresh THYE medium, and cells were grown to mid-logarithmic phase before being washed and resuspended in minimal medium (58) or in 1× PBS supplemented with 25 mM glucose. The membrane potential assays were conducted by measuring the florescence intensity of an anionic dye, DiSBAC1(3) [bis-(1,3-diethylthiobarbituric acid)trimethine oxonol, 5,5’-(prop-1-en-1-yl-3-ylidene)bis(2-tiobarbituric acid)], using the manufacturer's protocol for the FIVEphoton kit. As bis-oxonol is a lipophilic anionic molecule, the dye accumulates in the cell upon membrane depolarization, where it binds to intracellular components and results in an increase in cytosolic bis-oxonol fluorescence intensity. After incubating the cells with dye for 10 min, fluorescence was measured using the TECAN fluorescence plate reader (excitation wavelength of 530 nm and emission wavelength of 560 nm), and the fluorescence intensities were expressed as arbitrary units (AU). After dye stabilization, 1 mM CuSO4 was added. The plate conditions were set at 37°C with shaking for 5 s before every read, and fluorescence intensity measurements were taken every 15 min for at least 6 h. Carbonyl cyanide m-chlorophenylhydrazone (CCCP), a known membrane depolarizer, was used as a positive control, wherein the addition of 10 μM CCCP resulted in an immediate increase in fluorescence intensity. The change in membrane potential (ΔΨ, expressed as AU) was taken as the difference between the fluorescence value at a specific time point (every 15 min) of incubation (Ψf) and the initial stabilization value (Ψi). To control for the dye's intensity change, the value for the culture sample (Ψc) was normalized to the value for a blank with dye (Ψb). Statistical analysis was conducted using Student's t test, with a P value of <0.05 considered significant.
Genetic transformation assays.
Overnight cultures of S. mutans grown in THYE medium were pelleted, washed, diluted (1:20), and resuspended in chemically defined medium (43, 48). Cultures were allowed to grow at 37°C with 5% CO2 to mid-logarithmic growth phase (OD600 of ∼0.4). Cultures were then divided into three aliquots and treated with (i) no copper, (ii) 100 μM CuSO4, or (iii) 250 μM CuSO4. The copper concentrations used were based on the MICs calculated from assays in CDM: the MIC for S. mutans UA159 was 1,000 μM, and the MIC for the ΔcopYAZ strain was 500 μM CuSO4. Next, 1 μg of plasmid DNA (pDL277, containing a spectinomycin resistance marker) was added to 500-μl aliquots of cell suspensions and incubated in 5% CO2 at 37°C for 90 min. Following incubation, samples were briefly sonicated, serially diluted, and plated onto THYE agar with or without spectinomycin, to determine the number of transformants and the total number of possible viable cells, respectively. Transformation frequency was calculated by dividing the number of transformant CFU by the total number of viable CFU, times 100. Statistical analysis was conducted using Student's t test, with a P value of <0.05 considered significant.
RESULTS
Copper is toxic to S. mutans, and copYAZ is required for copper resistance in S. mutans.
To investigate the physiological function of the copYAZ operon in S. mutans, an isogenic ΔcopYAZ mutant was constructed and characterized in vitro. Growth kinetics and MICs were assessed using the wild-type and mutant strains under various concentrations of CuSO4 solutions. A 2-fold decrease in the MIC was observed in the ΔcopYAZ strain (2 mM CuSO4) compared with the MIC for the UA159 strain (4 mM CuSO4) under copper stress (P < 0.05). At higher concentrations (≥6 mM for the ΔcopYAZ strain and ≥12 mM for UA159), copper had a bactericidal effect on the viability of S. mutans. Complementation studies conducted with the CompΔcopYAZ (ΔcopYAZ harboring pIB-YAZ) strain showed restoration of copper-dependent growth inhibition to levels observed with the UA159 wild-type strain (MIC of 4 mM CuSO4; P < 0.05). Growth assays conducted with the wild-type, ΔcopYAZ, and CompΔcopYAZ strains revealed that exposure to added copper resulted in growth impairment of all strains and that loss of the copYAZ operon resulted in enhanced toxicity of copper relative to its toxicity for the wild-type and complemented strains (Fig. 1; see also Fig. S1 in the supplemental material). To validate that the effects observed in the growth assays were due to copper cations, the experiments were repeated with CuCl2, which showed an influence on the growth of S. mutans similar to that observed with CuSO4. In addition, we performed growth-inhibitory assays for the wild-type and ΔcopYAZ strains in the presence of various concentrations of other metal ions, including Ag, Cd, Zn, Mn, Mg, Hg, and Ca; the MICs from these assays did not reveal statistically relevant differences (data not shown). Hence, these results highlighted the importance and specificity of the CopYAZ system in copper-induced toxicity in S. mutans.
FIG 1.
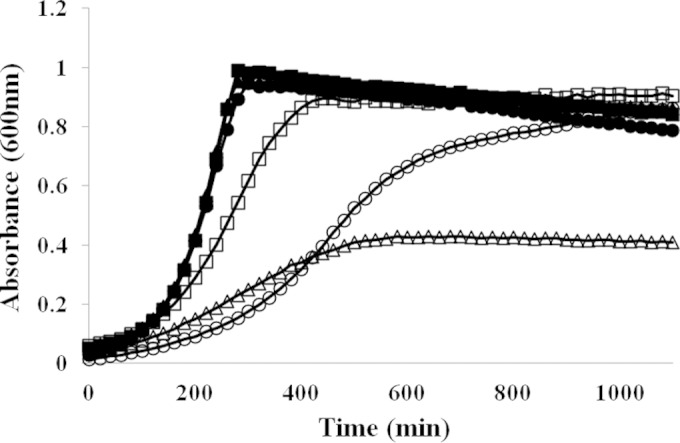
Growth analysis of S. mutans strains cultivated in the presence (open symbols) or absence (filled symbols) of 2 mM CuSO4. The results shown are representative of six independent experiments conducted with duplicates for each strain. Squares, S. mutans wild-type strain; triangles, ΔcopYAZ strain; circles, CompΔcopYAZ strain.
CopYAZ has a role in copper export in S. mutans.
In several Gram-positive bacteria, CopA functions in copper export. Copper transport studies using ICP-AES analysis revealed that S. mutans UA159 cells grown in the presence of 1 mM CuSO4 had a significantly large amount of intracellular copper (Fig. 2) compared to the amount in those grown without copper supplementation (no detectable intracellular copper). A 2-fold increase in the amount of intracellular copper was observed in the ΔcopYAZ strain compared with the amount in the wild-type strain (P < 0.05) (Fig. 2), thus suggesting the involvement of copYAZ in copper efflux in S. mutans. In the wild-type and ΔcopYAZ strains, transport studies conducted with Ag and Mn cations (using 50 μM AgNO3 or 1 mM MnCl2) showed no statistically significant differences in the intracellular levels of these cations (data not shown). The latter results support the specificity of the CopYAZ transport system for copper cations, which was further validated using the CopYAZ-complemented CompΔcopYAZ strain, which had an ability to efflux copper at levels comparable to the ability of the wild-type parent (Fig. 2). In the complementation experiments, we employed an E. coli strain deficient in copA (DW3110), which exhibits a defect in copper efflux and detoxification (59). The recombinant S. mutans copYAZ operon successfully complemented the deficiency in E. coli DW3110, as observed by a significantly decreased sensitivity to copper-induced killing and reduced intracellular copper ion accumulation (see Fig. S2A and B in the supplemental material), thus supporting a specific function for the S. mutans CopYAZ in copper transport and resistance.
FIG 2.
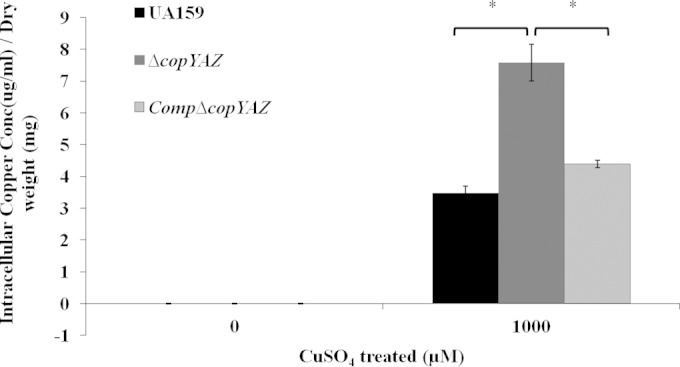
Role of CopYAZ in copper export. Cells of S. mutans wild-type strain UA159, the ΔcopYAZ strain, and the CompΔcopYAZ strain were grown in the presence or absence of 1 mM CuSO4 until the mid-logarithmic growth phase. Cells were lysed, and the intracellular copper concentrations, normalized to the cell dry weight, were determined. The results shown are the mean values obtained from three independent experiments ± standard errors. *, P < 0.05.
Copper inhibits biofilm formation, and copYAZ is required to tolerate copper stress under biofilm growth.
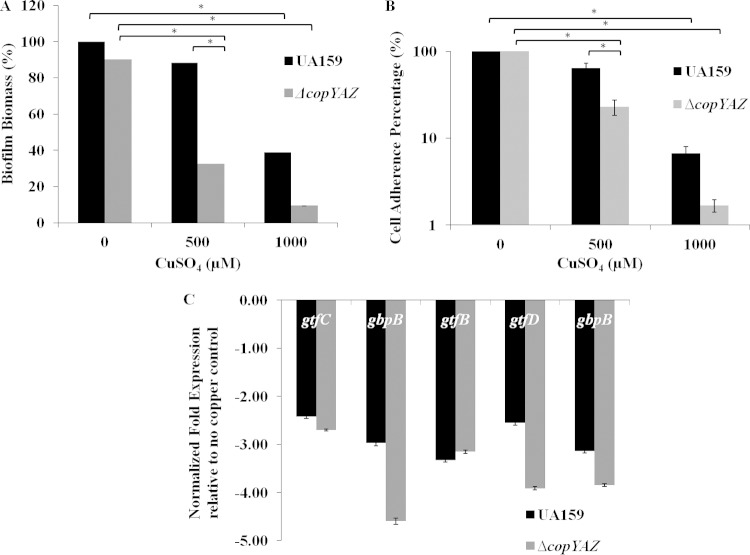
The wild-type and ΔcopYAZ strains produced comparable amounts of biomass in 18-h biofilms grown in the absence of copper. A significant reduction of biofilm biomass was observed in ΔcopYAZ strain biofilms cultivated with 500 μM CuSO4 relative to the biofilm biomass formed by the wild type (Fig. 3A). Biofilm biomass was drastically reduced at 1 mM CuSO4 in both wild-type and mutant strains (Fig. 3A). Initial cell attachment assays were conducted to evaluate the percentage of cells attached to a surface in the presence or absence of copper for different time intervals. At 1 h of incubation, the percentage of cells adhered in the presence of 500 μM CuSO4 was significantly reduced in both the wild-type (∼50% decrease) and the ΔcopYAZ strain (∼80% decrease) relative to the percentage in the no-copper control (Fig. 3B). At 1 mM CuSO4, a further reduction in the percentage of cells adhered was observed in both wild-type and the mutant strain. After prolonged incubation of 2 h or 4 h, no significant decrease in the number of cells adhering to the surface was observed in the wild-type strain at 500 μM CuSO4. However, theΔcopYAZ strain cultivated in the presence of copper showed significant reductions in the numbers of cells attached relative to the numbers for the no-copper control at all time periods (see Fig. S3 in the supplemental material). The results from these experiments showed that the addition of copper and deletion of the copYAZ operon reduced the ability of cells to adhere, likely leading to defective biofilms.
FIG 3.
Effects of copper on biofilm formation. (A) Biomass of 18-h biofilms derived from wild-type UA159 and the ΔcopYAZ strain grown in the absence or presence of CuSO4. The results shown represent the mean values obtained from three independent experiments with three replicates for each strain ± standard errors. *, P < 0.05. (B) UA159 and the ΔcopYAZ strain were incubated in the presence of various concentrations of CuSO4, and the percentages of cells adhered to the surface after 1 h of incubation relative to the adherence in the no-copper control were determined. The results shown represent the mean values obtained from three independent experiments with three replicates for each strain ± standard errors. *, P < 0.05. (C) Analysis of the expression of different biofilm matrix-related genes in UA159 and the ΔcopYAZ strain in the presence of 500 μM CuSO4 relative to their expression in the no-copper control. The results shown represent the mean values obtained from four independent experiments with three replicates for each strain ± standard errors. The P value was <0.05 for all the genes shown in the graph.
Copper affects the transcription of gtf and gbp genes.
To assess the underlying molecular mechanism associated with copper-mediated reduced cell adherence and reduced biofilm biomass of S. mutans, we examined the effects of copper on the transcription of gtf genes (gtfB, gtfC, and gtfD) and glucan binding protein (gbp) genes (gbpB and gbpC), whose products are needed to maintain the structural and functional integrity of the S. mutans biofilm (60–65). Gene expression analysis using qRT-PCR of biofilm-derived cDNAs with or without 500 μM CuSO4 demonstrated a significant, ≥2-fold downregulation of all five genes (P < 0.05) in the presence of copper relative to their transcription levels in the no-copper control (Fig. 3C). In the absence of copper, the transcription of gtf and gbp genes was not significantly affected in ΔcopYAZ and wild-type strain biofilms (data not shown). Furthermore, in the presence of copper, further transcriptional repression of gtfC, gtfD, gbpB, and gbpC was observed in ΔcopYAZ strain biofilms relative to their levels of transcription in the wild-type strain biofilms. In S. mutans, the negative regulatory role of copper in the transcription of the gbp genes, whose products facilitate biofilm cell adherence, as well as the transcription of the gtf genes, whose products are responsible for glucan production that maintains the integrity of the biofilm matrix, might explain the impaired biofilms observed under copper stress.
Copper induces oxidative stress, and CopYAZ modulates oxidative stress tolerance.
The copper-induced growth inhibition was assessed in the presence of the antioxidants glutathione and thiourea. The addition of 1.5 mM glutathione dramatically improved the growth of S. mutans UA159, ΔcopYAZ, and CompΔcopYAZ strains cultivated in the presence of 2 mM CuSO4 compared to the growth of the no-added-glutathione control (Fig. 4A). Treatment with glutathione in the absence of copper did not alter the growth of S. mutans strains used in this study (data not shown). The reversion of copper-induced growth defects was also observed in the presence of the antioxidant thiourea (data not shown). These results emphasize that copper-induced toxicity in S. mutans is likely dependent on copper-dependent generation of oxidative stress in S. mutans. Biofilms were also assessed to study the effect of glutathione (or thiourea) in reversal of the copper-induced biofilm defect by cultivating 18-h biofilms in the presence or absence of copper supplemented with or without 1.5 mM glutathione. Glutathione alone did not have any effect on the biofilm biomass of any of the three strains tested. The addition of 1.5 mM glutathione did not improve the biomass production of the biofilms cultivated in the presence of 500 μM or 1 mM copper in S. mutans UA159, ΔcopYAZ, and CompΔcopYAZ strains. These results suggest that S. mutans can utilize an alternate route to contend with copper stress during biofilm growth.
FIG 4.
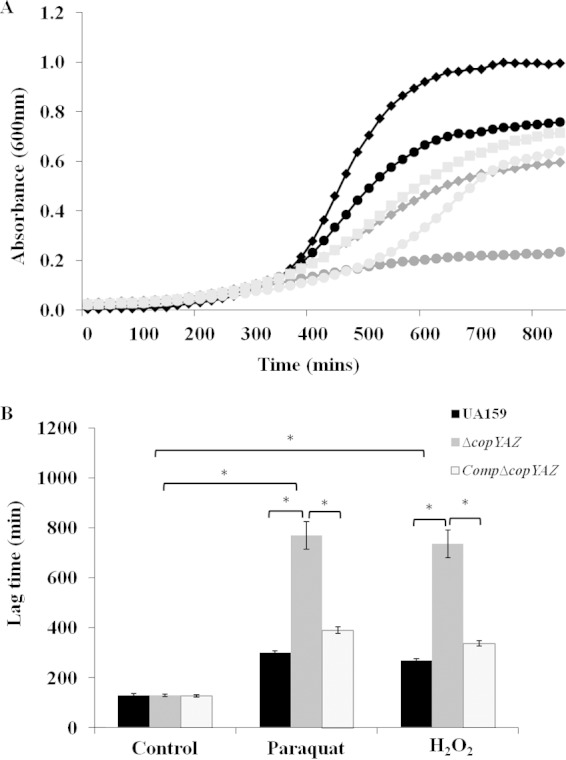
Copper-induced oxidative stress and involvement of CopYAZ in protection against oxidative stress. (A) Growth curves of wild-type UA159 (black), ΔcopYAZ (medium gray), and CompΔcopYAZ (light gray) strains in the presence of 2 mM CuSO4 copper with (diamonds and squares) or without (circles) 1.5 mM glutathione. The results shown are representative of six independent experiments conducted using duplicate samples for each strain. (B) Lag times for UA159, ΔcopYAZ, and CompΔcopYAZ strains to reach an OD600 of nearly 0.1 in the presence and absence of the oxidative stressors paraquat (25 mM) and H2O2 (0.0045%). The top two brackets indicate the results for statistical significance within strains under different stressors. The results are the means from three independent experiments conducted with three replicates for each strain. *, P < 0.001.
We next investigated the role of copYAZ in protecting the cells against oxidative stress. Growth kinetics analyses were performed using wild-type UA159, ΔcopYAZ, and CompΔcopYAZ strains in the presence of the oxidizers methyl viologen dichloride (paraquat) and hydrogen peroxide. Though all strains cultivated in the presence of either paraquat or hydrogen peroxide reached a comparable final optical density after 24 h (see Fig. S4 in the supplemental material), the lag phase of ΔcopYAZ was drastically prolonged relative to that of the wild type (Fig. 4B), thereby suggesting a role of the efflux system in protecting cells under oxidative stress. In the complemented strain CompΔcopYAZ, the lag phase in the presence of oxidative stressors was restored to a length similar to that of the wild type.
Copper inhibits growth under acid stress, and CopYAZ modulates the acid tolerance response of S. mutans.
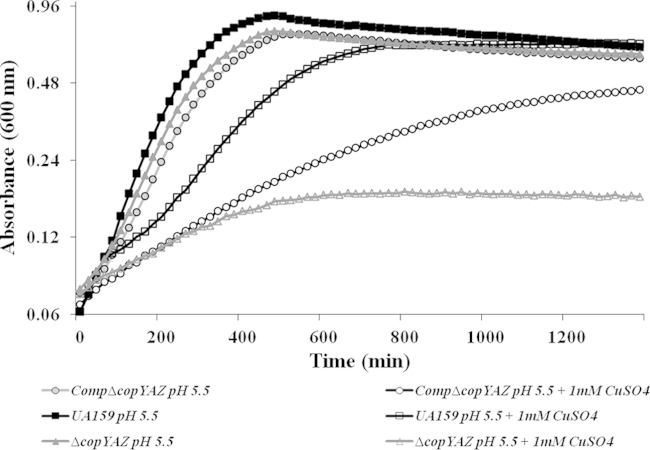
A previous report indicates that cuprous ions added to S. mutans strain GS-5 cultures can irreversibly inhibit the activity of F-ATPases, thereby compromising the ability to carry out glycolysis in acidic environments (27). Here, growth kinetic analyses were performed in the presence and absence of copper in neutral and acidic pH using wild-type UA159, ΔcopYAZ, and CompΔcopYAZ strains. Although the ΔcopYAZ strain displayed doubling times similar to those of the wild type under acid (pH 5.5) or copper (1 mM CuSO4) stress, the doubling times and the yield of the ΔcopYAZ strain were markedly impaired in THYE medium at pH 5.5 supplemented with 1 mM CuSO4 (Fig. 5). The combined effect of acid and copper stress caused approximately 2- and 5-fold increases in the doubling times of S. mutans UA159 and ΔcopYAZ strains, respectively, compared to their growth in the presence of either of these stressors alone (Fig. 5). These results emphasize the synergistic effects of copper and acid against the growth of S. mutans. The increased susceptibility of the ΔcopYAZ strain under acid and copper stress could be due to (i) surplus accumulation of copper ions or (ii) lack of cellular protection offered by the CopYAZ copper efflux system. To examine the importance of copYAZ in the acid adaptation process, we conducted acid tolerance response (ATR) assays. Not surprisingly, for all three strains tested, the percentage of adapted cells (previously subjected to pH 5.5) that survived was higher than that of nonadapted cells (maintained at pH 7.5). The loss of copYAZ decreased the ability of S. mutans to survive the pH 3.5 challenge relative to the survivability of the wild type, regardless of pre-acid adaptation, (Fig. 6). No significant difference in the percentages of surviving cells of the CompΔcopYAZ and wild-type strains at pH 3.5 was observed under either nonadapted or preadapted conditions, thereby demonstrating that copYAZ contributes to the ATR response in S. mutans.
FIG 5.
Growth of S. mutans under acid stress with or without copper. Growth curves of S. mutans UA159, ΔcopYAZ, and CompΔcopYAZ strains in the presence of acid (pH 5.5) and/or copper (1 mM CuSO4). The results shown are representative of six independent experiments with duplicate samples for each strain.
FIG 6.
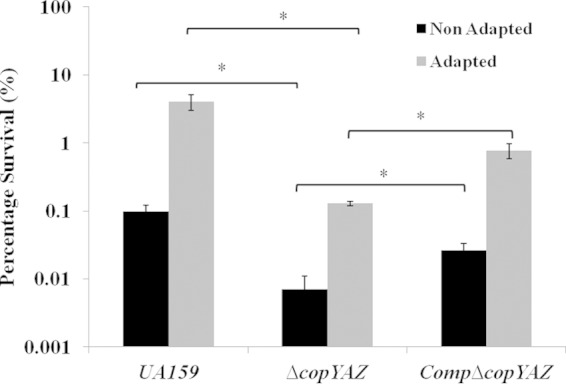
Acid tolerance response assays. Percentages of surviving S. mutans cells (in log scale) after 2 h of incubation at lethal pH of 3.5. Cells were either exposed (Adapted) or not exposed (Non Adapted) for preadaptation at the signal pH of 5.5. The results shown are the mean values obtained from three independent experiments ± standard errors. *, P < 0.05.
Previously, copper has been shown to reduce the activity of F-ATPase in S. mutans (27). Therefore, we examined the role of copper in the transcription of acid stress-related genes that include components of the S. mutans F0F1 ATPase (SMU.1530 [atpD], SMU.1531 [atpE], SMU.1532 [atpF], and SMU.1534 [atpH]) and uvrA, a DNA damage repair gene that is induced under acid stress (66). The expression levels of these genes in the wild-type strain cultivated in the presence and absence of copper were not significantly different. However, in the ΔcopYAZ strain, the transcription of uvrA was downregulated significantly, by more than 2-fold (P < 0.05) and 3.6-fold (P < 0.05) for incubation with and without copper, respectively, relative to the transcription levels in the wild-type strain. A significant, 2.2-fold reduction (P < 0.05) in the expression of atpH was observed in the ΔcopYAZ strain in the presence of copper relative to its expression in the control without copper. Studies are under way to analyze the effects of copper and the copYAZ operon on the acid-inducible regulon of S. mutans that is responsible for the observed acid-sensitive phenotype in this oral pathogen.
Copper induces membrane depolarization, and CopYAZ helps maintain membrane potential.
In E. coli and Salmonella, contact killing on copper surfaces involves immediate membrane depolarization, leading to compromised cell viability (11). To test whether copper and CopYAZ modulate the membrane potential of S. mutans, we conducted fluorometric assays with bis-oxonol dyes, which can enter depolarized cells and bind intracellular proteins or membranes. Higher fluorescence intensities, as a result of elevated influx of the dye, indicate increased membrane depolarization. In S. mutans, the addition of copper initiated a gradual increase in the fluorescence intensity, which was maximal relative to that in the no-copper control after 15 min of incubation (Fig. 7A). The increase in fluorescence intensity suggested that copper influx induced a dissipation of the membrane potential. The S. mutans ΔcopYAZ cells exhibited a depolarized membrane phenotype where the cells cultivated even in the absence of copper showed a gradual and sustained increase in the DiSBAC1(3)-dependent fluorescence intensity compared with the fluorescence intensity in the wild type (Fig. 7B). Furthermore, the restoration of the fluorescence intensity in the complemented strain CompΔcopYAZ to the wild-type level provided additional evidence for the involvement of this operon in the membrane depolarization of S. mutans. In membrane potential assays, CCCP was used as a positive control; the addition of 10 μM CCCP resulted in an immediate increase in fluorescence intensity as a result of instant membrane depolarization (data not shown).
FIG 7.
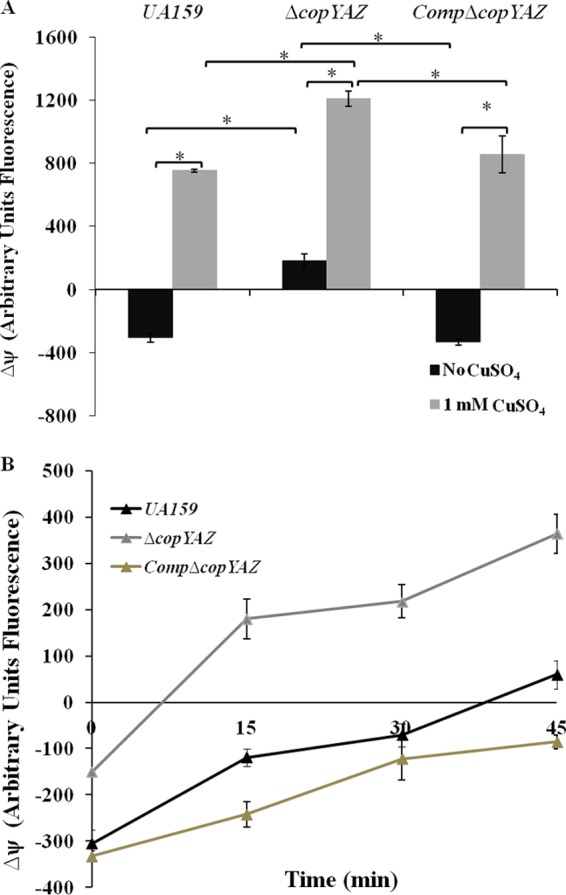
Membrane potential in cells exposed to copper. The changes in membrane potential (ΔΨ) of S. mutans strains over 45 min were measured using a bis-oxonol probe. (A) Fluorescence intensities of cells after 15 min of incubation in the absence or presence of 1 mM CuSO4. The statistical significance of the results at each time point was calculated using Student's t test. *, P < 0.05. (B) Fluorescence intensities of wild-type UA159, ΔcopYAZ, and CompΔcopYAZ strains in the absence of copper over a period of 45 min. Data shown are the mean results from four independent experiments ± standard errors. Statistically significant differences between the results for the ΔcopYAZ strain and the other two strains were observed at each time point. P < 0.05.
The addition of copper and loss of copYAZ alter the transformability of S. mutans.
Genetic transformation assays were conducted to investigate the effect of copper on the genetic competence development of S. mutans. These assays were conducted in two different growth media, THYE medium and CDM, which activate separate competence induction pathways via the CSP-activated ComDE and XIP-activated ComRS signaling pathway, respectively (43, 67). When UA159 and the ΔcopYAZ strain were grown in the peptone-abundant, nutrient-rich THYE medium supplemented with 10 μM, 100 μM, 250 μM, 500 μM, or 1 mM CuSO4, a 5-fold decrease in transformation frequency relative to the frequency in the controls without copper was observed in the wild-type strain only at the 1 mM concentration of CuSO4 (P < 0.05; data not shown). In the ΔcopYAZ mutant, transformability was significantly reduced, by over 10-fold relative to that of the wild type, irrespective of copper supplementation (data not shown). The effects of copper on transformability were stronger when cells were grown in CDM. For instance, in CDM, a 20-fold reduction in transformation frequency was observed in both the wild-type and ΔcopYAZ strains in the presence of 100 μM CuSO4 relative to the frequency in the no-copper control (P < 0.05) (Fig. 8). Furthermore, in the presence of copper, we noted that the transformability of wild-type and mutant strains was significantly reduced in a dose-dependent manner (Fig. 8); our comparison of the numbers of viable recipient cells in these strains showed that total cell viability in UA159 and ΔcopYAZ strains was not affected at these copper concentrations (see Fig. S6 in the supplemental material). Also of interest, loss of the copper efflux system in the ΔcopYAZ strain impaired its transformability by over 30-fold relative to that of the wild type (P < 0.05), even in the absence of copper stress (Fig. 8). To argue against the possibility of this transformation defect arising due to copper-mediated plasmid DNA damage, we conducted transformation assays using cells preincubated with copper. These cells were washed to remove exogenous or cell-associated copper and then supplemented with fresh medium containing plasmid DNA. A similar decrease in genetic transformation was observed in the presence of copper relative to the transformation frequency of the no-copper control, thereby confirming that the defect in genetic transformation was indeed due to copper present within the cells (data not shown).
FIG 8.
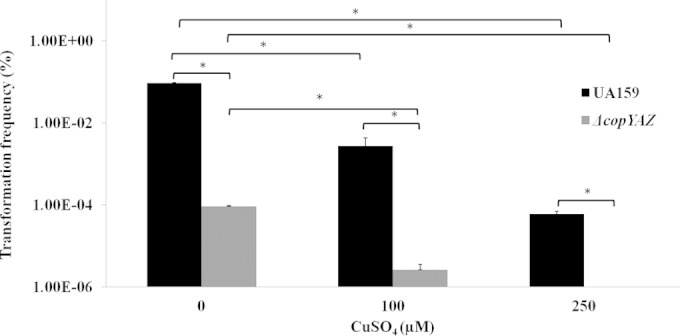
Transformation frequencies (in log scale) for S. mutans UA159 and the ΔcopYAZ strain in the presence of various concentrations of copper. Frequencies were calculated as the number of transformant CFU divided by the total number of viable CFU, times 100. The results are the means from three independent experiments conducted in triplicates. *, P < 0.05.
Copper represses the expression of genes associated with genetic competence.
To determine the pathways by which copper and CopYAZ modulate the transformability of S. mutans, we compared the expression levels of the competence-related genes comC, comD, comE, comR, comS, and comX in the isogenic strains treated with or without copper. In S. mutans UA159, supplementation with 100 μM CuSO4 reduced the expression of comX and comS by 2-fold and 2.8-fold, respectively, compared with the expression levels in the no-copper control (P < 0.001). Regardless of the presence of copper, the expression of comX, which is critical for competence development, was reduced by 2.5-fold in the ΔcopYAZ strain compared with its expression in the wild-type strain (P < 0.001). Moreover, relative to the expression in the wild type, the expression of comS in the copper transporter mutant was repressed more than 8-fold and 12.5-fold in the absence and presence of copper, respectively (P < 0.001).
DISCUSSION
Fluctuations in intracellular levels of copper can have severe implications for the physiology of all prokaryotes. The mechanisms involved in copper-induced killing vary among bacterial species. Bacteria usually initiate a global adaptive genetic response to copper, depending on their physiological copper requirements and the presence of copper in their environmental niches (68–73). While in some bacteria, such as Enterococcus faecalis and Pseudomonas aeruginosa, a large portion of the genome (approximately 300 genes) is differentially expressed under copper stress, in others, such as Lactococcus lactis, only 11 genes are copper responsive (70, 72, 73). While we did not analyze changes in the entire copper regulon of S. mutans, targeted gene expression analysis in this work revealed that copper-responsive genes included the gtfBCD and gbpBC genes, which are associated with biofilm formation, the comS and comX genes, which are critical for competence development, and the uvrA and atpH genes, which are involved in acid stress tolerance. An important aspect of this work was also to validate the role of the S. mutans CopYAZ system in copper efflux, which influenced the bacterium's ability to tolerate environmental stressors and maintain its membrane potential. The link between copper homeostasis via the CopYAZ system and its effects on comS and comX transcription for competence development is novel. Since the competence pathway is closely linked with the biofilm and stress tolerance pathways of S. mutans (38, 46, 47, 74), this work adds to the current knowledge as to how these pathways can be modulated by environmental copper stress.
The results of growth kinetic assays and MIC testing in this work indicated that higher concentrations of copper are toxic to S. mutans and that copper toxicity is counteracted through CopYAZ activity. In most bacteria, CopA has been speculated or proven to remove copper cations from the cell (30, 31, 59), which we validated using copper transport assays. Since we observed that cells of both the wild-type and the ΔcopYAZ strain were able to accumulate copper cations, we suspect that in S. mutans, copper import/influx occurs passively, in a nonspecific manner, as demonstrated in other bacteria, such as Helicobacter pylori and E. coli (75–77).
In the oral cavity, since S. mutans is constantly exposed to fluctuating levels of oxidative stress, its ability to adapt to such conditions is important for its survival. Here, we demonstrate that the addition of glutathione dramatically reversed the copper-induced growth defect in S. mutans cultures, thus suggesting that the toxicity of copper is likely a result of copper-dependent generation of oxidative stress in S. mutans. Copper can generate oxidative stress either by depleting glutathione (due to its capability of forming a complex with glutathione) (78) or by generating reactive oxygen species (ROS), especially superoxide radicals, which are toxic to cells (3, 30). Copper, due to its high reactivity, is capable of replacing Fe or Mn in the active site of proteins, thereby rendering them inactive (79). In S. mutans, the proteins encoded by perR (peroxide regulator), sod (superoxide dismutase), and dpr (dps-like peroxidase resistance protein) are associated with resistance to oxidative stress (80–83). The dpr-encoded protein requires Fe for its activity, the sod-encoded protein requires Mn/Fe, and PerR has been speculated to require Mn or Fe (80, 82–84). Hence, it can be speculated that copper replaces Fe/Mn in the active sites of the products of dpr, sod, and/or perR, thereby inactivating these proteins and compromising the cell viability under oxidative stress. However, further research is warranted to dissect the specific underlying mechanisms involved in this process. Our results also demonstrated the importance of the S. mutans copYAZ operon in resistance against oxidative stress, thereby suggesting its involvement in the stress adaptation process of this oral pathogen.
Copper and acidic pH exerted a synergistic effect against the growth of S. mutans. Growth in THYE medium at pH 5.5 or in THYE medium with 1 mM CuSO4 did not produce discernibly different phenotypes in the ΔcopYAZ and UA159 strains. However, significant increases in the doubling times were noted when these strains were cultivated in THYE medium at pH 5.5 with 1 mM CuSO4, thus suggesting a synergistic effect of acid and copper stress against the growth of S. mutans. Since cuprous ions in S. mutans have previously been associated with impaired glycolysis under acid stress (27), our study supports this finding by demonstrating a notable growth defect in the presence of copper and acid stress. The impaired growth observed for the ΔcopYAZ strain in the presence of copper and acid stress could be due to the accumulation of surplus copper in the cells or to the involvement of the copYAZ operon in protection under acid stress. S. mutans produces lactic acid as a metabolic end product of dietary sugars and mounts an ATR that affords it growth and survival under pH values as low as 3.5 (53, 85). During the ATR, S. mutans undergoes a number of physiological changes, such as increased synthesis of stress-responsive proteins, membrane fatty acid changes, and increased activity of proton-translocating ATPases (53, 85). Such adaptation requires preexposure to a sublethal signal of pH 5.5 to activate the processes to protect cells against killing pH values (pH 2.0 to 3.0) (53, 85). Severe decreases in the viability of both adapted and nonadapted cells of the ΔcopYAZ strain compared with those of the wild type at pH 3.5 implicated the contribution of copYAZ to the ATR of S. mutans. In the presence of copper, the transcriptional repression of the acid stress-related genes uvrA and atpH likely contributed to the observed acid-sensitive phenotype of S. mutans. The repression of the expression of uvrA in the ΔcopYAZ strain also supported the association of the copYAZ operon with the acid stress response. Although we did not analyze the effect of copper in influencing the acid-inducible regulon of S. mutans, the results of this study offer a foundation for understanding the copper-mediated acid tolerance response in this oral pathogen. Further studies are under way to elucidate the effect of copper in modulating the proteome of S. mutans, with the specific aim of determining the proteins involved in the acid adaptation process under copper stress.
Under normal growth conditions, bacteria maintain their membrane potential by establishing multiple ion gradients across their intact cytoplasmic membrane. The maintenance of intact cell membranes is crucial to maintain ATP hydrolysis and proton motive force (86, 87). A disturbance in the amount of ions can result in hyperpolarization (higher negative intracellular electrical potential) or depolarization (higher positive intracellular electrical potential) of the membrane potential. A rational target for copper-induced toxicity in bacteria is their cell wall or cell membrane, as these are the parts of bacteria that are exposed to external stress. Reports have indicated that, in Gram-positive bacteria, the metal binding sites usually lie within the peptidoglycan layer (88, 89). Copper also tends to accumulate on the inner side of the cell membrane, making the inner side more positive and, thus, causing membrane depolarization (11, 90). Here, we demonstrated the effect of copper on initiating an immediate membrane potential dissipation, which was significantly higher in the ΔcopYAZ strain than in the wild-type strain. The S. mutans ΔcopYAZ cells exhibited a depolarized membrane phenotype, even in the absence of copper, compared with the membrane phenotype of wild-type cells. The gradual increase in membrane depolarization observed in cells of the ΔcopYAZ strain could be due to membrane perturbation caused by the loss of the CopA transmembrane protein. Our findings are in agreement with those of another study, where the loss of the arsenic and antimony efflux system in E. coli was shown to be linked with cell membrane depolarization (91). Disturbances in the bacterial membrane potential can alter certain cellular processes, such as cell division and differentiation, maintenance of the integrity of cellular membranes, electron transport across the membranes, localization of specific proteins and protein complexes, and regulation of the levels of cellular energy (86, 92–94). In S. mutans, copper acting as a membrane potential dissipater might contribute to the generation of copper-dependent oxidative and acid stress, where both these stresses are likely to be affected by variations in the bacterial membrane integrity and/or changes in electron transport across the bacterial membrane (87, 95–97).
Copper has an inhibitory effect on biofilm formation in S. mutans. This mechanism relies on the ability of copper to inhibit early attachment of cells to the surface and to significantly reduce biomass production in S. mutans. The reduction in cell adherence and biomass production was more prominent in the ΔcopYAZ strain, likely due to surplus accumulation of copper within the cells. In the presence of sucrose, cell wall-associated Gtf proteins mediate the synthesis of d-glucose polysaccharides, which are glucans that promote S. mutans cell adherence to the tooth surface and to other adhered bacteria (62, 65). S. mutans produces three different Gtf proteins: GtfB, which produces water-insoluble glucans composed predominantly of α-1,3 linkages; GtfD, which mostly produces soluble α-1,6-linked glucans; and GtfC, which synthesizes both α-1,3 and α-1,6 glucans (98–100). In our experiments, the expression levels of gtfB, gtfC, and gtfD were significantly repressed by copper. The transcriptional repression of these gtf genes validated the effects of copper in influencing genes critical for glucan production and biofilm matrix formation; their downregulation under conditions of excess copper explains the poor biofilm biomass that we observed. In addition to Gtf proteins, surface-associated Gbp proteins in S. mutans promote cell-cell aggregation by mediating the binding of S. mutans to glucans (61, 101, 102). The addition of copper significantly reduced the transcription of gbpB and gbpC, further expanding the knowledge of components affected by environmental copper. The downregulation of these gbp genes distinctly emphasized the effect of copper in the initial sucrose-dependent biofilm formation, where expression of these genes is essential for the transition from planktonic to biofilm growth (102, 103). In contrast to our gtf transcription results, others showed previously that supplementation with 1 mM copper induced the transcription and translation of gtfD but not gtfBC (28). In another study, RNAs isolated from planktonic cells were used for expression analysis (31), whereas this study utilized RNAs from 18-h biofilms cultivated with or without added copper. In the absence of copper, we did not note any significant differences in the transcription levels of gtfBCD and gbpBC between the ΔcopYAZ and wild-type strains; hence, it is not surprising that we obtained comparable amounts of biofilm biomass for these strains. In addition to the repressive effects of copper on gtfBCD and gbpBC transcription, loss of copper efflux in the ΔcopYAZ mutant led to reduced expression of the gtf and gbp genes in cop mutant biofilms with added copper in the medium. These results provide strong evidence that copper is extremely effective in inhibiting biofilm formation, one of the most vital virulence attributes of this oral pathogen.
The influence of copper and the CopYAZ system on the development of genetic competence and the expression of comX and comS in S. mutans is novel. The ComDE and ComRS signaling pathways involved in S. mutans competence development differ in their mechanisms of action (42–45). The CSP-ComDE pathway is activated by CSP, whose precursor peptide is encoded by comC (38, 44, 104). Upon activation of ComD and subsequent phosphorylation, transfer to ComE leads to the activation of ComX, which is a critical switch required for the competent state of S. mutans (44, 47). Recently, the proximal regulator of ComX was determined to be ComR, an Rgg-like transcriptional regulator in the cytosol that is able, in conjunction with internalized XIP, to activate ComX for the transcription of late competence genes required for DNA uptake and recombination (43). It has been shown that the nutrients and peptones present in the growth medium of S. mutans can differentially affect the CSP and XIP activities needed for competence activation (42, 43, 45, 67, 105). CDM was shown to be optimal only for XIP-mediated competence induction, whereas the peptone-rich THYE medium was optimal for transformation in the presence of CSP (42, 43, 45, 67). In our assays, since the effect of copper on S. mutans transformability is dramatically pronounced in CDM relative to the effect in THYE medium, we speculate that its influence on genetic competence occurs primarily via the XIP-induced signaling pathway. Moreover, the observation that only comS and comX out of all competence-related genes tested (i.e., comCDE, comRS, and comX) had significantly changed expression under our test conditions provides additional evidence that the effects of copper on transformation are modulated by the XIP-ComRS pathway. The fact that copper affects the expression of comS but not comR suggests that copper represses the activity of ComX by affecting the function or secretion of ComS and/or XIP, thus causing a dramatic reduction in transformation frequency. In the absence of copper, a significant reduction in the transformation frequency of the ΔcopYAZ strain relative to that of the wild type implies the importance of this operon in genetic competence development in S. mutans. Previously, in Streptococcus pneumoniae, inactivation of a putative metal-transporting operon (AdcCBA) was shown to confer a competence-deficient phenotype (106, 107). The operon was speculated to be involved in zinc transport, and the addition of zinc improved the transformability of the adc null mutants (106, 107). In our study, although the mechanism involved in modulation of competence by the copYAZ system is not fully understood, two suggestions can be made based on the observed transcriptional repression of comS: (i) the CopYAZ may have a role in modulating the function, processing, or export of XIP, and/or (ii) the membrane perturbation that results from the loss of CopA may hinder the process of DNA acquisition, resulting in reduced transformation frequency.
In conclusion, the results of the present study enhance our understanding about the effects of copper on S. mutans survival under planktonic and biofilm growth conditions. We also demonstrate the role of the copYAZ system in maintaining S. mutans physiology under stress conditions. Insight into the mechanism of copper toxicity and its connection with biofilm formation and genetic transformation is instrumental in understanding and devising new strategies to utilize copper as an effective antibiofilm agent to combat S. mutans infections. Since S. mutans can be considered an ideal model organism to study the genetics and physiology of pathogenic Gram-positive bacteria (108), our study holds relevance in suggesting the importance of copper acquisition and homeostasis in closely related pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the technical assistance provided by Kirsten Krastel in performing the MIC assays and DNA cloning for this study.
This research was supported by NIH RO1DE013230-03 and CIHR-MT15431 grants to D.G.C. C.M.L. is the recipient of a Canada Research Chair. K.S. is the recipient of Cell Signaling Fellowship CIHR-STP-53877.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02433-14.
REFERENCES
- 1.Samanovic MI, Ding C, Thiele Dennis J, Darwin KH. 2012. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe 11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreini C, Banci L, Bertini I, Rosato A. 2008. Occurrence of copper proteins through the three domains of life: a bioinformatic approach. J Proteome Res 7:209–216. doi: 10.1021/pr070480u. [DOI] [PubMed] [Google Scholar]
- 3.Rademacher C, Masepohl B. 2012. Copper-responsive gene regulation in bacteria. Microbiology 158(Pt 10):2451–2464. doi: 10.1099/mic.0.058487-0. [DOI] [PubMed] [Google Scholar]
- 4.Dupont CL, Grass G, Rensing C. 2011. Copper toxicity and the origin of bacterial resistance—new insights and applications. Metallomics 3:1109–1118. doi: 10.1039/c1mt00107h. [DOI] [PubMed] [Google Scholar]
- 5.Arredondo M, Núñez MT. 2005. Iron and copper metabolism. Mol Aspects Med 26:313–327. doi: 10.1016/j.mam.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung DKC, Lau WY, Chan WT, Yan A. 2013. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J Bacteriol 195:4556–4568. doi: 10.1128/JB.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rensing C, Franke McDevitt S. 2013. The copper metallome in prokaryotic cells, p 417–450. In Banci L. (ed), Metal ions in life sciences, vol 12 Metallomics and the cell. Springer, Dordrecht, Netherlands. [DOI] [PubMed] [Google Scholar]
- 10.Achard MES, Tree JJ, Holden JA, Simpfendorfer KR, Wijburg OL, Strugnell RA, Schembri MA, Sweet MJ, Jennings MP, McEwan AG. 2010. The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence. Infect Immun 78:2312–2319. doi: 10.1128/IAI.01208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol 14:1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 12.Hodgkinson V, Petris MJ. 2012. Copper homeostasis at the host-pathogen interface. J Biol Chem 287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsh PD. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 14.Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C. 2005. The global burden of oral diseases and risks to oral health. Bull World Health Organ 83:661–669. [PMC free article] [PubMed] [Google Scholar]
- 15.Dreizen S, Spies HA, Spies TD. 1952. The copper and cobalt levels of human saliva and dental caries activity. J Dent Res 31:137–142. doi: 10.1177/00220345520310011001. [DOI] [PubMed] [Google Scholar]
- 16.Grytten J, Tollefsen T, Afseth J. 1987. The effect of a combination of copper and hexetidine on plaque formation and the amount of copper retained by dental plaque bacteria. Acta Odontol Scand 45:429–433. [DOI] [PubMed] [Google Scholar]
- 17.Grytten J, Aamdal Scheie A, Afseth J. 1988. Effect of a combination of copper and hexetidine on the acidogenicity and copper accumulation in dental plaque in vivo. Caries Res 22:371–374. [DOI] [PubMed] [Google Scholar]
- 18.Drake DR, Grigsby W, Cardenzana A, Dunkerson D. 1993. Synergistic, growth-inhibitory effects of chlorhexidine and copper combinations on Streptococcus mutans, Actinomyces viscosus, and Actinomyces naeslundii. J Dent Res 72:524–528. [DOI] [PubMed] [Google Scholar]
- 19.Mahler DB. 1997. The high-copper dental amalgam alloys. J Dent Res 76:537–541. [DOI] [PubMed] [Google Scholar]
- 20.Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol Rev 50:353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamada S, Slade HD. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev 44:331–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afseth J, Amsbaugh SM, Monell-Torrens E, Bowen WH, Rølla G, Brunelle J, Dahl E. 1984. Effect of copper applied topically or in drinking water on experimental caries in rats. Caries Res 18:434–439. doi: 10.1159/000260799. [DOI] [PubMed] [Google Scholar]
- 23.Afseth J. 1983. Some aspects of the dynamics of Cu and Zn retained in plaque as related to their effect on plaque pH. Scand J Dent Res 91:169–174. [DOI] [PubMed] [Google Scholar]
- 24.Bales CW, Freeland-Graves JH, Askey S, Behmardi F, Pobocik RS, Fickel JJ, Greenlee P. 1990. Zinc, magnesium, copper, and protein concentrations in human saliva: age- and sex-related differences. Am J Clin Nutr 51:462–469. [DOI] [PubMed] [Google Scholar]
- 25.Afseth J, Oppermann RV, Rølla G. 1983. Accumulation of Cu and Zn in human dental plaque in vivo. Caries Res 17:310–314. doi: 10.1159/000260682. [DOI] [PubMed] [Google Scholar]
- 26.Maltz M, Emilson C-G. 1988. Effect of copper fluoride and copper sulfate on dental plaque, Streptococcus mutans and caries in hamsters. Scand J Dent Res 96:390–392. [DOI] [PubMed] [Google Scholar]
- 27.Dunning JC, Ma Y, Marquis RE. 1998. Anaerobic killing of oral streptococci by reduced, transition metal cations. Appl Environ Microbiol 64:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P-M, Chen J-Y, Chia J-S. 2006. Differential regulation of Streptococcus mutans gtfBCD genes in response to copper ions. Arch Microbiol 185:127–135. doi: 10.1007/s00203-005-0076-2. [DOI] [PubMed] [Google Scholar]
- 29.Ridge PG, Zhang Y, Gladyshev VN. 2008. Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One 3:e1378. doi: 10.1371/journal.pone.0001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solioz M, Abicht H, Mermod M, Mancini S. 2010. Response of Gram-positive bacteria to copper stress. J Biol Inorg Chem 15:3–14. doi: 10.1007/s00775-009-0588-3. [DOI] [PubMed] [Google Scholar]
- 31.Odermatt A, Suter H, Krapf R, Solioz M. 1993. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem 268:12775–12779. [PubMed] [Google Scholar]
- 32.Lu ZH, Solioz M. 2001. Copper-induced proteolysis of the CopZ copper chaperone of Enterococcus hirae. J Biol Chem 276:47822–47827. doi: 10.1074/jbc.M106218200. [DOI] [PubMed] [Google Scholar]
- 33.Solioz M, Stoyanov JV. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev 27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 34.Vats N, Lee SF. 2001. Characterization of a copper-transport operon, copYAZ, from Streptococcus mutans. Microbiology 147:653–662. [DOI] [PubMed] [Google Scholar]
- 35.Page WJ, Sadoff HL. 1976. Physiological factors affecting transformation of Azotobacter vinelandii. J Bacteriol 125:1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trombe M-C. 1993. Characterization of a calcium porter of Streptococcus pneumoniae involved in calcium regulation of growth and competence. J Gen Microbiol 139:433–439. doi: 10.1099/00221287-139-3-433. [DOI] [PubMed] [Google Scholar]
- 37.Page WJ, Doran JL. 1981. Recovery of competence in calcium-limited Azotobacter vinelandii. J Bacteriol 146:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y-H, Lau PCY, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cvitkovitch DG. 2001. Genetic competence and transformation in oral streptococci. Crit Rev Oral Biol Med 12:217–243. doi: 10.1177/10454411010120030201. [DOI] [PubMed] [Google Scholar]
- 40.Johnsborg O, Eldholm V, Håvarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol 158:767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Federle MJ, Morrison DA. 2012. One if by land, two if by sea: signalling to the ranks with CSP and XIP. Mol Microbiol 86:241–245. doi: 10.1111/mmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MØ, Thiede B, Petersen FC. 2012. Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J Bacteriol 194:3781–3788. doi: 10.1128/JB.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y-H, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. 2012. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett 336:104–112. doi: 10.1111/j.1574-6968.2012.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen FC, Tao L, Scheie AA. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol 187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aspiras MB, Ellen RP, Cvitkovitch DG. 2004. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol Lett 238:167–174. doi: 10.1111/j.1574-6968.2004.tb09752.x. [DOI] [PubMed] [Google Scholar]
- 48.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suntharalingam P, Senadheera MD, Mair RW, Lévesque CM, Cvitkovitch DG. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol 191:2973–2984. doi: 10.1128/JB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49:193–205. doi: 10.1016/S0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 51.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. doi: 10.1099/mic.0.2008/019265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perry JA, Lévesque CM, Suntharaligam P, Mair RW, Bu M, Cline RT, Peterson SN, Cvitkovitch DG. 2008. Involvement of Streptococcus mutans regulator RR11 in oxidative stress response during biofilm growth and in the development of genetic competence. Lett Appl Microbiol 47:439–444. doi: 10.1111/j.1472-765X.2008.02455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Senadheera D, Krastel K, Mair R, Persadmehr A, Abranches J, Burne RA, Cvitkovitch DG. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J Bacteriol 191:6415–6424. doi: 10.1128/JB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarrete JU, Borrok DM, Viveros M, Ellzey JT. 2011. Copper isotope fractionation during surface adsorption and intracellular incorporation by bacteria. Geochim Cosmochim Acta 75:784–799. doi: 10.1016/j.gca.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senadheera MD, Lee AWC, Hung DCI, Spatafora GA, Goodman SD, Cvitkovitch DG. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J Bacteriol 189:1451–1458. doi: 10.1128/JB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujiwara S, Kobayashi S, Nakayama H. 1978. Development of a minimal medium for Streptococcus mutans. Arch Oral Biol 23:601–602. doi: 10.1016/0003-9969(78)90280-7. [DOI] [PubMed] [Google Scholar]
- 59.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci U S A 97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowen WH, Koo H. 2011. Biology of Streptococcus mutans derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res 45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA. 2007. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol Lett 268:158–165. doi: 10.1111/j.1574-6968.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol 192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakano K, Matsumura M, Kawaguchi M, Fujiwara T, Sobue S, Nakagawa I, Hamada S, Ooshima T. 2002. Attenuation of glucan-binding protein C reduces the cariogenicity of Streptococcus mutans: analysis of strains isolated from human blood. J Dent Res 81:376–379. doi: 10.1177/154405910208100604. [DOI] [PubMed] [Google Scholar]
- 64.Mattos-Graner RO, Porter KA, Smith DJ, Hosogi Y, Duncan MJ. 2006. Functional analysis of glucan binding protein B from Streptococcus mutans. J Bacteriol 188:3813–3825. doi: 10.1128/JB.01845-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun 61:3811–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanna MN, Ferguson RJ, Li Y-H, Cvitkovitch DG. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J Bacteriol 183:5964–5973. doi: 10.1128/JB.183.20.5964-5973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol 194:3774–3780. doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baker J, Sitthisak S, Sengupta M, Johnson M, Jayaswal RK, Morrissey JA. 2010. Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl Environ Microbiol 76:150–160. doi: 10.1128/AEM.02268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kershaw CJ, Brown NL, Constantinidou C, Patel MD, Hobman JL. 2005. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151:1187–1198. doi: 10.1099/mic.0.27650-0. [DOI] [PubMed] [Google Scholar]
- 70.Teitzel GM, Geddie A, De Long SK, Kirisits MJ, Whiteley M, Parsek MR. 2006. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa. J Bacteriol 188:7242–7256. doi: 10.1128/JB.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, Morrissey JA. 2011. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol 81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 72.Magnani D, Barré O, Gerber SD, Solioz M. 2008. Characterization of the CopR regulon of Lactococcus lactis IL1403. J Bacteriol 190:536–545. doi: 10.1128/JB.01481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abrantes M, Lopes Mde F, Kok J. 2011. Impact of manganese, copper and zinc ions on the transcriptome of the nosocomial pathogen Enterococcus faecalis V583. PLoS One 6:e26519. doi: 10.1371/journal.pone.0026519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahn S-J, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rensing C, Grass G. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- 76.Cooksey DA. 1993. Copper uptake and resistance in bacteria. Mol Microbiol 7:1–5. doi: 10.1111/j.1365-2958.1993.tb01091.x. [DOI] [PubMed] [Google Scholar]
- 77.Waidner B, Melchers K, Stähler FN, Kist M, Bereswill S. 2005. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J Bacteriol 187:4683–4688. doi: 10.1128/JB.187.13.4683-4688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Helbig K, Bleuel C, Krauss GJ, Nies DH. 2008. Glutathione and transition-metal homeostasis in Escherichia coli. J Bacteriol 190:5431–5438. doi: 10.1128/JB.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Irving H, Williams RJP. 1953. The stability of transition-metal complexes. J Chem Soc (Resumed) 0:3192–3210. doi: 10.1039/JR9530003192. [DOI] [Google Scholar]
- 80.Fujishima K, Kawada-Matsuo M, Oogai Y, Tokuda M, Torii M, Komatsuzawa H. 2013. dpr and sod in Streptococcus mutans are involved in coexistence with S sanguinis, and PerR is associated with resistance to H2O2. Appl Environ Microbiol 79:1436–1443. doi: 10.1128/AEM.03306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamamoto Y, Higuchi M, Poole LB, Kamio Y. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J Bacteriol 182:3740–3747. doi: 10.1128/JB.182.13.3740-3747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamamoto Y, Poole LB, Hantgan RR, Kamio Y. 2002. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J Bacteriol 184:2931–2939. doi: 10.1128/JB.184.11.2931-2939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin ME, Byers BR, Olson MO, Salin ML, Arceneaux JE, Tolbert C. 1986. A Streptococcus mutans superoxide dismutase that is active with either manganese or iron as a cofactor. J Biol Chem 261:9361–9367. [PubMed] [Google Scholar]
- 84.Nakayama K. 1992. Nucleotide sequence of Streptococcus mutans superoxide dismutase gene and isolation of insertion mutants. J Bacteriol 174:4928–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matsui R, Cvitkovitch D. 2010. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol 5:403–417. doi: 10.2217/fmb.09.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strahl H, Hamoen LW. 2010. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A 107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bakker EP, Mangerich WE. 1981. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J Bacteriol 147:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beveridge TJ, Murray RG. 1980. Sites of metal deposition in the cell wall of Bacillus subtilis. J Bacteriol 141:876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beveridge TJ, Forsberg CW, Doyle RJ. 1982. Major sites of metal binding in Bacillus licheniformis walls. J Bacteriol 150:1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li F, Feterl M, Warner JM, Keene FR, Collins JG. 2013. Dinuclear polypyridylruthenium(II) complexes: flow cytometry studies of their accumulation in bacteria and the effect on the bacterial membrane. J Antimicrob Chemother 68:2825–2833. doi: 10.1093/jac/dkt279. [DOI] [PubMed] [Google Scholar]
- 91.Meng Y-L, Liu Z, Rosen BP. 2004. As(III) and Sb(III) Uptake by GlpF and Efflux by ArsB in Escherichia coli. J Biol Chem 279:18334–18341. doi: 10.1074/jbc.M400037200. [DOI] [PubMed] [Google Scholar]
- 92.Barak I, Muchova K. 2013. The role of lipid domains in bacterial cell processes. Int J Mol Sci 14:4050–4065. doi: 10.3390/ijms14024050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Busch KB, Deckers-Hebestreit G, Hanke GT, Mulkidjanian AY. 2013. Dynamics of bioenergetic microcompartments. Biol Chem 394:163–188. doi: 10.1515/hsz-2012-0254. [DOI] [PubMed] [Google Scholar]
- 94.Ramamurthi KS, Losick R. 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A 106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marquis RE. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol 15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- 96.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 97.Dimroth P, Kaim G, Matthey U. 2000. Crucial role of the membrane potential for ATP synthesis by F(1)F(o) ATP synthases. J Exp Biol 203:51–59. [DOI] [PubMed] [Google Scholar]
- 98.Hanada N, Kuramitsu HK. 1988. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect Immun 56:1999–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanada N, Kuramitsu HK. 1989. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect Immun 57:2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gilpin ML, Russell RR, Morrissey P. 1985. Cloning and expression of two Streptococcus mutans glucosyltransferases in Escherichia coli K-12. Infect Immun 49:414–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schilling KM, Bowen WH. 1992. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect Immun 60:284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duque C, Stipp RN, Wang B, Smith DJ, Höfling JF, Kuramitsu HK, Duncan MJ, Mattos-Graner RO. 2011. Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans. Infect Immun 79:786–796. doi: 10.1128/IAI.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Welin J, Wilkins JC, Beighton D, Svensäter G. 2004. Protein expression by Streptococcus mutans during initial stage of biofilm formation. Appl Environ Microbiol 70:3736–3741. doi: 10.1128/AEM.70.6.3736-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Son M, Ahn S-J, Guo Q, Burne RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dintilhac A, Alloing G, Granadel C, Claverys J-P. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol 25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 107.Dintilhac A, Claverys JP. 1997. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol 148:119–131. [DOI] [PubMed] [Google Scholar]
- 108.Lemos JA, Quivey RG, Koo H, Abranches J. 2013. Streptococcus mutans: a new Gram-positive paradigm? Microbiology 159(Pt 3):436–445. doi: 10.1099/mic.0.066134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.