FIG 1.
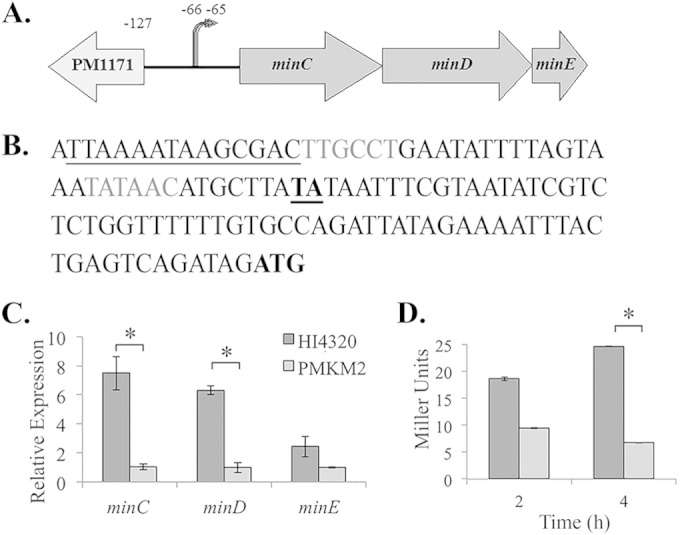
The min operon is positively regulated by RcsB. (A) Organization of the minCDE open reading frame. minC (702 bp), minD (813 bp), and minE (207 bp) are located in the same operon. The noncoding region upstream of minC is 127 bp long. Two transcriptional start sites for minCDE are located at positions −65 and −66 upstream of the minC start codon. (B) Sequence upstream of minCDE. The start codon of minC is in boldface. The two transcriptional start sites identified using 5′-RACE are underlined and in boldface. They are 65 and 66 bp upstream of the start codon. Upstream of the two identified start sites are putative −10 and −35 regions (gray letters). A potential RcsB binding site is underlined based on the E. coli consensus sequence (TAAGAATAATCCTA). (C) qRT-PCR analysis of minC, minD, and minE expression levels in P. mirabilis HI4320 and P. mirabilis HI4320 rcsB::Strr (PMKM2). minC expression is 7.5 times lower in PMKM2 than in the wild type, minD expression is 6.3 times lower in PMKM2 than in the wild type, and minE expression is 2.4 times lower in PMKM2 than in the wild type. (D) β-Galactosidase activity of pMinCLacZ in the PM7002 wild type (PMKH6) (dark-gray bars) and PM7002 rcsB::Strr (PMKH7) (light-gray bars). The activity of the minC promoter is decreased 2-fold in PMKH7 compared to PMKH6 after 2 h on solid agar and is decreased 4-fold in PMKH7 after 4 h on solid agar. The asterisks indicate a P value of <0.05.
