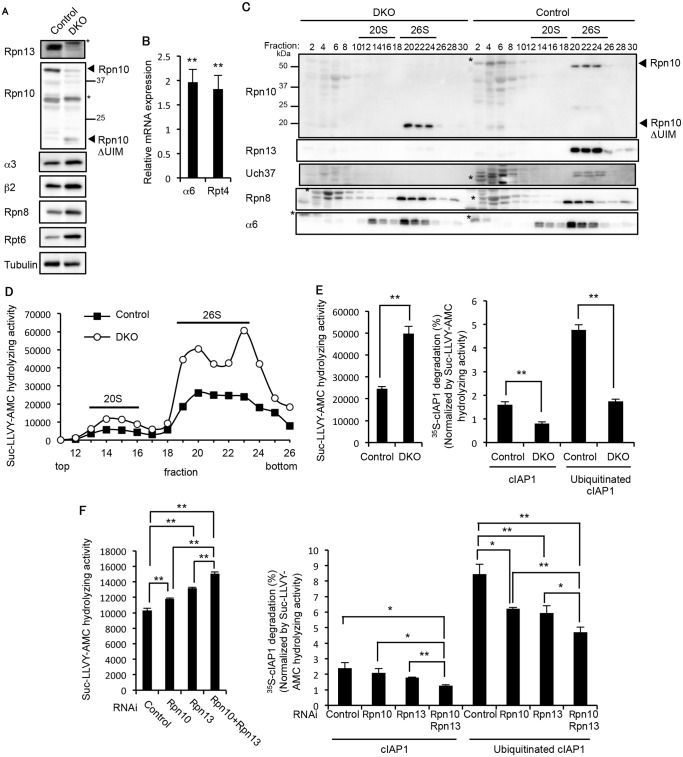
Fig 4. Redundant roles of Rpn10 and Rpn13 in degradation of ubiquitinated proteins.
(A) Immunoblot analysis of liver lysates from 3-week-old control and DKO mice with antibodies against the indicated proteins. (B) Real-time RT-PCR was performed to measure the mRNA expressions of the proteasome subunits α6 and Rpt4 in the liver of 2–4-week-old control and DKO mice. Data represent transcript levels in DKO livers relative to those in control livers and are expressed as means; error bars denote SEM. **p < 0.01 (n = 7 for each genotype). (C) Lysates from control and DKO livers were fractionated by glycerol gradient centrifugation (8 to 32% glycerol from fraction 1 to 30) and an equal amount of each fraction was used for immunoblot analysis using antibodies against the indicated proteins. Asterisks indicate nonspecific bands. (D) Each fraction of (C) was assayed for chymotrypsin-like activity using Suc-LLVY-AMC as a substrate. (E) The 26S proteasome fractions of (D) (fractions 20–23) were subjected to the assay of chymotrypsin-like activity (left panel). Degradation rates of 35S-labeled cIAP1 with or without ubiquitination were measured and normalized by chymotrypsin-like activity (right panel). Data are mean ± standard deviations from triplicate experiments. **p < 0.01. (F) Lysates from HeLa cells transfected with indicated siRNAs were assayed for Suc-LLVY-AMC hydrolyzing activity (left panel) and degradation of 35S-labeled cIAP1 with or without ubiquitination. cIAP degradation rates are normalized by Suc-LLVY-AMC hydrolyzing activity (right panel). Data are mean ± standard deviations from three experiments. *p < 0.05; **p < 0.01.