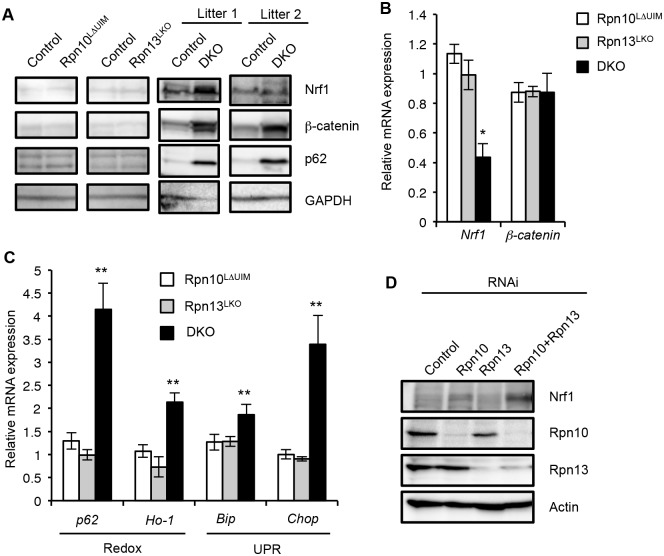
Fig 6. Synthetic effect of Rpn10-UIM and Rpn13 deletion on degradation of ubiquitinated proteins and cellular stress.
(A) Immunoblot analysis of whole-cell extracts of livers from indicated genotypes of mice (2-month-old for Rpn10LΔUIM and Rpn13LKO, and 2–4-week-old DKO) with antibodies against indicated proteins. (B and C) Real-time RT-PCR was used to measure the expression of transcripts encoding Nrf1, β-catenin, redox pathway (p62 [18412] and Ho-1 [15368]), and UPR pathway (Bip [14828] and Chop [13198]) genes in the livers of 3–6-week-old control and DKO mice. Data represent levels of transcripts in each genotype liver relative to those in control liver and are expressed as means; error bars denote SEM. *p < 0.05; **p < 0.01 (n = 3 for Rpn10LΔUIM and Rpn13LKO, and n = 4 for DKO). (D) HeLa cells were transfected with siRNA against Rpn10 and Rpn13. Where indicated, cells were transfected with a mixture of siRNAs. After 96h, whole-cell extracts were subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies.