Fig 2. Algorithm for discrete TI model.
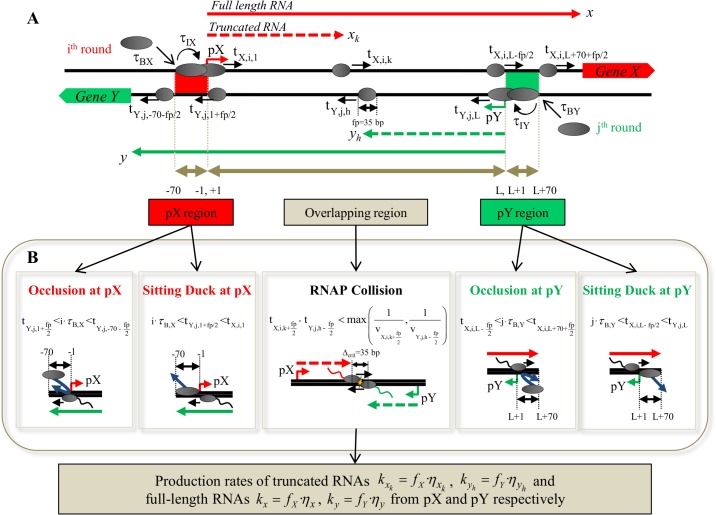
(A) A pair of convergent promoters pX (present on sense/top DNA strand) and pY (present on antisense/bottom DNA strand) separated by overlapping DNA of length L is shown. Promoters pX and pY drive expression of genes X and Y respectively, which produce full-length transcripts x and y (denoted by bold arrows) respectively. For each ith and jth round of transcription from pX and pY promoters respectively, RNAP (denoted by large grey ovals) form DNA-bound RNAP complexes at the respective promoter region following a binding (τBX and τBY) and initiation (τIX and τIY) process. After firing, the center of RNAP moves to first position of the overlapping region to form an elongation complex (EC, denoted by smaller grey ovals). The time taken for each ith EC (fired from pX) to reach kth position on sense strand (tX,i,k) as well as the time taken for each jth EC (fired from pY) to reach hth position on the antisense (tY,j,h) strand along the overlapping DNA are tracked. The footprint of an EC is denoted by fp. (B) For each ith and jth rounds of transcription, the model calculates the outcome of TI depending on the region where opposing RNAPs meet. Occlusion and sitting duck interference occur at the promoters pX (left panels) or pY (right panels), and RNAP collisions between ECs occur along the overlapping DNA (middle panel) following the mathematical constraints shown. Upon RNAP collision, one or both ECs on the sense and antisense strand fall off the DNA and result in production of truncated transcripts x k and y h (denoted by dashed arrows) from pX and pY respectively. In absence of any kind of TI, transcription is successful, producing a full-length transcript (x, y). Once 30,000 rounds of transcription from the stronger promoter have been calculated the net rate of production of full-length (k x and k y) and truncated RNA ( and ) are obtained.