Abstract
Mitogen-activated protein kinase (MAPK) cascades are critical signaling modules that mediate the transduction of extracellular stimuli into intracellular response. A relatively large number of MAPKKKs have been identified in a variety of plant genomes but only a few of them have been studied for their biological function. In the present study, we identified an Arabidopsis Raf-like MAPKKK gene Raf43 and studied its function in biotic and abiotic stress response using a T-DNA insertion mutant raf43-1 and two Raf43-overexpressing lines Raf43-OE#1 and Raf43-OE#13. Expression of Raf43 was induced by multiple abiotic and biotic stresses including treatments with drought, mannitol and oxidative stress or defense signaling molecule salicylic acid and infection with necrotrophic fungal pathogen Botrytis cinerea. Seed germination and seedling root growth of raf43-1 were significantly inhibited on MS medium containing mannitol, NaCl, H2O2 or methyl viologen (MV) while seed germination and seedling root growth of the Raf43-OE#1 and Raf43-OE#13 lines was similar to wild type Col-0 under the above stress conditions. Soil-grown raf43-1 plants exhibited reduced tolerance to MV, drought and salt stress. Abscisic acid inhibited significantly seed germination and seedling root growth of the raf43-1 line but had no effect on the two Raf43-overexpressing lines. Expression of stress-responsive RD17 and DREB2A genes was significantly down-regulated in raf43-1 plants. However, the raf43-1 and Raf43-overexpressing plants showed similar disease phenotype to the wild type plants after infection with B. cinerea or Pseudomonas syringae pv. tomato DC3000. Our results demonstrate that Raf43, encoding for a Raf-like MAPKKK, is required for tolerance to multiple abiotic stresses in Arabidopsis.
Introduction
Unlike animals, plants are sessile and cannot avoid unfavorable environmental stresses by moving to a different location; however, they have evolved a set of complicated mechanisms to timely sense and effectively respond to abiotic and biotic stresses such as drought, high salinity and extreme temperatures as well as pathogen infection and herbivore infestation [1–3]. Sensing and recognition of extracellular environmental stimuli and invading pathogens through surface/extracellular receptors such as receptor-like kinases and receptor-like proteins often activate complex downstream signaling networks, which ultimately initiate effective intracellular responses [2]. During these signaling processes, mitogen-activated protein kinase (MAPK) cascades play important roles in relaying and amplifying stimulus-specific signals to the cellular machinery through modifying a set of specific target proteins via the way of phosphorylation [4–7].
The MAPK cascade is highly conserved across different species and comprises three functional protein kinases, i.e. MAPK kinase kinases (MAPKKKs), MAPK kinases (MAPKKs) and MAPKs [4]. Compared with relatively small numbers of the MAPK and MAPKK families (e.g. 20 MAPKs and 10 MAPKKs in Arabidopsis) [8, 9], MAPKKKs form a larger family that contain more than 70 members in Arabidopsis, rice, maize, tomato, canola and Gossypium raimondii [10–15]. MAPKKKs act biochemically and functionally at the top of MAPK cascades but they have more variable structures and domain compositions when compared with MAPKs and MAPKKs. Based on the sequences in the kinase catalytic domain, the plant MAPKKKs are divided into three subfamilies, namely, MEKK, Raf and ZIK [10] or three groups (Group A, B and C) [8]. There are 21 MEKKs, 48 Rafs and 11 ZIKs among 80 MAPKKKs in Arabidopsis [10].
Compared with the extensive studies exploring the biological functions for the MAPKs and MAPKKs, functional characterization of MAPKKKs is relatively lagging. Recent studies using loss-of-function and gain-of-function mutants have demonstrated that MAPKKKs play important roles in growth and development as well as in abiotic and biotic stress response. In Arabidopsis, at least four MAPKKKs belonging to the MEKK subfamily have been shown to be involved in regulating growth and development, e.g. ANP1, ANP2 and ANP3 regulate cell division while YODA controls stomatal development [16–18]. The Arabidopsis MEKK1 forms MEKK1–MKK1/2–MPK4 cascade that negatively regulates defense responses against biotrophic pathogens but positively regulates defenses against necrotrophic fungi and is also involved in stress response and diverse development process [19–24]. Several MAPKKKs from other plants such as tomato MAPKKKα and MAPKKKε, tobacco NPK1, Nicotiana benthamiana NbMAPKKKα, NbMAPKKKβ and NbMAPKKKγ and alfalfa OMTK1 have also been implicated in regulating immune response, programmed cell death and development process [25–30]). More recently, it was found that a RXLR effector PexRD2 from Phytophthora infestans could target to N. benthamiana MAPKKKε to perturb plant immunity-related signaling [31].
Among the MAPKKK families characterized in plants at genome-wide level so far, approximately 60% of the members belong to the Raf subfamily; for instances, 48 Rafs out of total 80 MAPKKKs in Arabidopsis [10], 45 of 75 in rice [11], 46 of 74 in maize [12] and 39 of 66 in canola [14]. However, only a small fraction of the Raf-like MAPKKKs have been identified and studied at molecular and genetic levels for their biological functions. The biological importance of the Raf-like MAPKKKs was highlighted by the molecular identification and detailed functional analyses of Constitutive Triple Response 1 (CTR1) and ENHANCED DISEASE RESISTENCE1 (EDR1) in Arabidopsis [32, 33]. CTR1 is a Raf-like MAPKKK that modulates a negative regulation of ethylene pathway by acting upstream of MKK9-MPK3/6 proteins [34–36] and is also involved in sugar response [37]. EDR1, another Raf-like MAPKKK, functions at the top of a MAPK cascade to regulate negatively the salicylic acid (SA)-inducible defense responses [33] and directly associates with MKK4/MKK5 for negatively regulating innate immunity [38]. The Arabidopsis SIS8/AT6 was recently reported to play roles in salt stress and sugar responses [39, 40], whereas MAP3Kδ4 was shown to regulate growth and shoot branching [41]. In rice, a Raf-like MAPKKK DSM1 is involved in drought tolerance [42] while another Raf-like MAPKKK ILA1 regulates mechanical tissue formation in the leaf lamina joint [43]. The rice EDR1 was found to negatively regulate rice bacterial resistance via activation of ethylene biosynthesis [44]. However, the functions of most other Raf-like MAPKKKs in plants are as yet unknown.
In bioinformatics analysis of publicly available microarray data to identify putative stress-responsive MAPKKKs in Arabidopsis, we found that a gene, Raf43, encoding for a Raf-like MAPKKK, could be induced by drought, salt and oxidative stress, pathogen infection and elicitor treatment. In the present study, we performed functional analyses using a T-DNA insertion mutant line raf43-1 and two Raf43-overexpresing lines Raf43-OE#1 and Raf43-OE#13 to explore the possible roles of Raf43 in abiotic and biotic stress response. Our results showed that Raf43 could be induced by multiple abiotic and biotic stresses and that the raf43-1 mutant plants exhibited reduced tolerance to osmotic, drought, salt and oxidative stresses while the Raf43-overexpressing plants did not show any altered stress response. The raf43-1 mutant showed increased sensitivity to abscisic acid (ABA) and the raf43-1 plants constitutively down-regulated the expression of stress-responsive gene. However, the raf43-1 mutant and the Raf43-overexpressing plants showed similar disease phenotype to the wild type plants in response to infection with necrotrophic fungal pathogen B. cinerea or bacterial pathogen Pseudomonas syringae pv. tomato DC3000. These findings demonstrate that Raf43, encoding for a Raf-like MAPKKK, is required for tolerance to multiple abiotic stresses in Arabidopsis.
Material and Methods
Plant growth and treatments
Arabidopsis plants were grown in a mixture of perlite: vermiculite: plant ash with a ratio of 1:6:2, respectively, under fluorescent light (200 μE·m2·s-1) at 22°C with 60% relative humidity. Seed dormancy was broken by soaking the seeds in water for 2 days in a 4°C refrigerator and then used for experiments. Four-week-old plants (ecotype Col-0) were used for different treatments to analyze the expression of Raf43 in response to stresses. For drought treatment, rosette leaves were cut and placed on laboratory bench under similar conditions as control intact plants. For mannitol treatment, plants were irrigated with 350 mM mannitol or water as a control. For treatment with methyl viologen (MV) and salicylic acid (SA), solutions of 50 μM MV, 1 mM SA or same volume of water as a control were sprayed onto the plants. Inoculation of B. cinerea was conducted as below. Leaf samples were taken at indicated time points after treatment/inoculation and stored at -80°C until use.
Characterization of raf43-1 mutant
A T-DNA insertion line FLAG_505A06 for Raf43 in Ws-0 background was obtained from Arabidopsis thaliana Resource Centre for Genomics at the Versailles Genetics and Plant Breeding Laboratory, France. A three-primer PCR-based genotyping strategy was conducted using gene-specific primers FLAG_505A06-LP (GCG CAG TTG ATT ATA CGA GAA G) and FLAG_505A06-RP (GCG AGG AGG AGT ACT GTG ATG) with a T-DNA primer F-LB4 (CGT GTG CCA GGT GCC CAC GGA ATA GT) to identify wild type, heterozygous and homozygous individuals. Plants homozygous for the Raf43 mutation were used for further experiments and this line was designated as raf43-1.
Generation of Raf43-overexpressing and lines
For construction of overexpression vector, a 1450 bp fragment of raf43 gene was amplified using a pair of primers Raf43-1F (ATG GAT GGA GAG GTT ACT TCT TGG A) and Raf43-1R (CAC AAA AAT GAG TGA CTC TAG ACG T) and cloned into pUC19 by T/A cloning, yielding a recombinant plasmid pUC19-Raf43. After confirmation by sequencing, the open reading frame (ORF) of Raf43 was amplified using a pair of primers Raf43-2F (AGT GGA TCC ATG GAT GGA GAG GTT ACT TCT, a BamHI site underlined) and Raf43-2R (GCA GTC GAC TCA AGC GAA TTT AGG CTT AGGT, a SalI site underlined) with the recombinant plasmid pUC19-Raf43 as template and cloned into binary vector pCAMBIA 99–1, resulting a recombinant plasmid pCAMBIA991-Raf43. For construction of complementation vector, the Raf34 ORF was amplified from pUC19-Raf43 using primers Raf43-2F and Raf43-2R and cloned into pCAMBIA 1301 at BamHI/SalI sites, yielding plasmid pCAMBIA1301-Raf43. A 1295 bp fragment upstream the start codon of the Raf43 gene was amplified from genomic DNA using a pair of primers pRaf43-1F (AGT GAATTC AGCAGGGAGAGGGATAGGGT, a EcoRI site underlined) and pRaf43-1R (AGT GGATCC GTTTAAACCCCTCCCCAAAAC, a BamHI site underlined) and cloned into pCAMBIA1301-Raf43 at EcoRI/BamHI sites, yielding plasmid pCAMBIA1301-proRaf43-Raf43. Plasmids pCAMBIA991-Raf43 and pCAMBIA1301-proRaf43-Raf43 were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation using GENE PULSER II Electroporation System (Bio-Rad Laboratories, Hercules, CA, USA). Arabidopsis transformation was performed using the floral dip method [45]. The overexpression plasmid pCAMBIA991-Raf43 was introduced into Col-0 background while the complementation plasmid pCAMBIA1301-proRaf43-Raf43 was introduced into raf43 background (harboring Basta resistance selection marker). Seeds from transformed plants (T0) were harvested and sown on 1/2 MS supplemented with 50 μg/mL hygromycin (Hgr). Seedlings of the T1 generation were selected and self-pollinated. T1 progenies on selective medium with Hgr-resistant/Hgr-sensitive segregating ratio of 3:1 were selected and transformed to soil for self-pollination. Progenies of the individual T1 plants were observed on selective medium and those showing 100% resistance to Hgr were selected as homozygous lines. Homozygous overexpression lines Raf43-OE#1 and Raf43-OE#13 and homozygous complementation lines Raf43-C1 and Raf43-C17 were used for further studies.
Abiotic stress assays
In seed germination and root growth assays, seeds were surface-sterilized and plated on 1/2 MS medium supplemented with or without different concentrations of mannitol, NaCl, MV, H2O2, or ABA and incubated at 4°C for 2 days to synchronize germination. The plates were placed in a growth chamber under long day condition and germination was counted at 7 days after incubation. Root growth of the seedlings was measured after placing the Petri dishes vertically for 10 days. In whole plant abiotic stress assays, three-week-old soil-grown plants were used. Drought stress was applied to plants by withholding water for 7–10 days while salt stress was applied to plants by drenching with solution of 200 mM NaCl. Oxidative stress treatment was conducted by foliar spraying with 50 mM MV onto leaves of the plants. After a period of treatment, the numbers of plants that continued to grow were recorded and the survival rate was calculated. In these abiotic stress assays, plants grown under normal condition or without abiotic stress treatment were used as controls. Chlorophyll content of leaf tissues of the plants in salt and oxidative stress assays were measured according to the method described previously [46]. Chlorophyll content was calculated according to the formula Chl (A+B) = 5.24A664+22.24A648, where Chl is the chlorophyll concentration in micrograms per milliliter and A is the absorption. Relative water content (RWC) in leaves of the plants in drought stress assays was measured according to the method described previously [47]. Fully expanded leaves were detached from 6 individual plants to measure the leaf fresh weight (WF), turgid leaf weight (WT), and dry weights (WD) and RWC were calculated from the equation RWC (%) = (WF-WD)/(WT-WD)×100% [47].
Disease Assays
Disease assays with B. cinerea were performed according to previously reported procedure [46]. B. cinerea was grown on 2 × V8 agar (36% V8 juice, 0.2% CaCO3 and 2% agar) at 22°C for 10–12 days. Spores were collected by dissolving in 1% maltose media and filtered through bi-layered cheesecloth. Spore density in spore suspension was adjusted to 1×105 spores/mL. In detached leaf inoculation assays, fully expanded leaves from at least 10 individual plants were inoculated by dropping a 5 μL of spore suspension onto leaf surface. In whole plant inoculation assays, 4-week-old plants were inoculated by foliar spraying with spore suspension or buffer (as a mock-inoculation control). The inoculated leaves and plants were kept in sealed trays or tanks with high relative humidity at 22°C to facilitate disease development. The diameter of each lesion in detached leaf inoculation assays was measured at 5 days after inoculation.
P. syringae was cultivated on plates of King’s B (KB) medium supplemented with 50 mg/mL rifampicin and resuspended into 10 mM MgCl2 at OD600 = 0.002 for plant inoculation. Four-week-old plants were infiltrated at two sites of leaves with 20 μL aliquots of the bacterial suspension using 1-ml syringes without needles. After inoculation, plants were covered with a transparent plastic film and disease symptoms were observed daily. For measurement of bacterial titers in inoculated leaves, leaf punches (6 mm in diameter) were collected at 0, 2 or 4 days post infiltration, surface sterilized in 70% ethanol for 10 seconds, homogenized in 200 μL of 10 mM MgCl2, diluted in 10 mM MgCl2, and plated on KB agar plates containing rifampicin at 100 μg/mL.
Analyses of gene expression
For Genevestigator-based analyses, microarray expression data from various datasets were obtained using GENEVESTIGATOR (https://www.genevestigator.com/gv/). The expression data for Raf43 (Locus At3g46930) under different abiotic and biotic stress conditions were mined. Most of the expression data were mined from ecotype Col-0 except for a set of the expression data in P. syringae pv. tomato DC3000 experiments was obtained from ecotype Ws-0. Basically, only expression changes of Raf43 in stress/elicitor-treated or pathogen-infected plants >2 folds over controls with p values <0.05 were used. The microarray expression analyses were obtained from samples treated with cold, drought, osmotic, oxidative or salt stresses, infected by pathogens such as Alternaria brassicicola (Ab), B. cinerea (Bc), Blumeria graminis f. sp. hordei (Bg), P. syringae pv. maculicola (Psm), P. syringae pv. phaseolicola (Psp), P. syringae pv. tomato (Pst) DC3000, or Sclerotinia sclerotiorum (Ss) or treated with elicitor including fungal chitin, bacterial EF-Tu (elf18 and elf26), FLG22 and hrpZ, Arabidopsis Pep2, and chemical inducer SA. For qRT-PCR-based analyses, total RNA was extracted from leaf samples using TRIZOL reagent (Invitrogen, Shanghai, China) and treated using DNase RQ1 (Promega, Madison, USA) to remove contaminant genomic DNA. First-strand cDNA was reversely transcribed using AMV reverse transcriptase (Takara, Dalian, China) with oligod(T) primer according to the manufacturer’s instructions. Primers used in qRT-PCR-based analyses of gene expression were Raf43-3F, TCG TCG TCC CGC AAA GAT; Raf43-3R, TTC CCG TGA GCA AAC CTA; AtABA2-F, CTC GCT TTG GCT CAT TTG C; AtABA2-R, CCG TCA GTT CCA CCC CTT T; AtNCED3-F, CCG GTG GTT TAC GAC AAG AA; AtNCED3-R, CCC AAG CGT TCC AGA GAT G; AtRD17-1F, ACG TCC ACG CCG TTG GT; AtRD17-1R, CTC CGG ATG TTC CAC TGG AA; AtDREB2A-1F, GAC CTA AAT GGC GAC GAT GT, AtDREB2A-1R, TCG AGC TGA AAC GGA GGT AT; AtUBC-1F: TCA AAT GGA CCG CTC TTA TC; AtUBC-1R, CAC AGA CTG AAG CGT CCA AG. qRT-PCR was conducted with SYBR premix Ex Taq (Takara, China) in a CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The expression levels were quantified using 2−ΔΔCT method. The data obtained from qRT-PCR were normalized using the levels of the UBC gene and the gene expression levels in raf43-1, Raf43-overexpressing or treated/inoculated plants were shown as folds of the levels in the control untreated or mock-inoculated plants, which were set as 1.0.
Statistical analysis
All experiments were repeated independently for three times and at least 10 plants were included in each treatment in an independent experiment. Data obtained from three biological replications were subjected to statistical analysis by the Student’s t-test method using DPS software (http://www.chinadps.net/index.htm) and the results are presented as mean ± standard errors. The probability values of p<0.05 were considered as significant between different treatments.
Results
Raf43 is responsive to multiple abiotic and biotic stresses
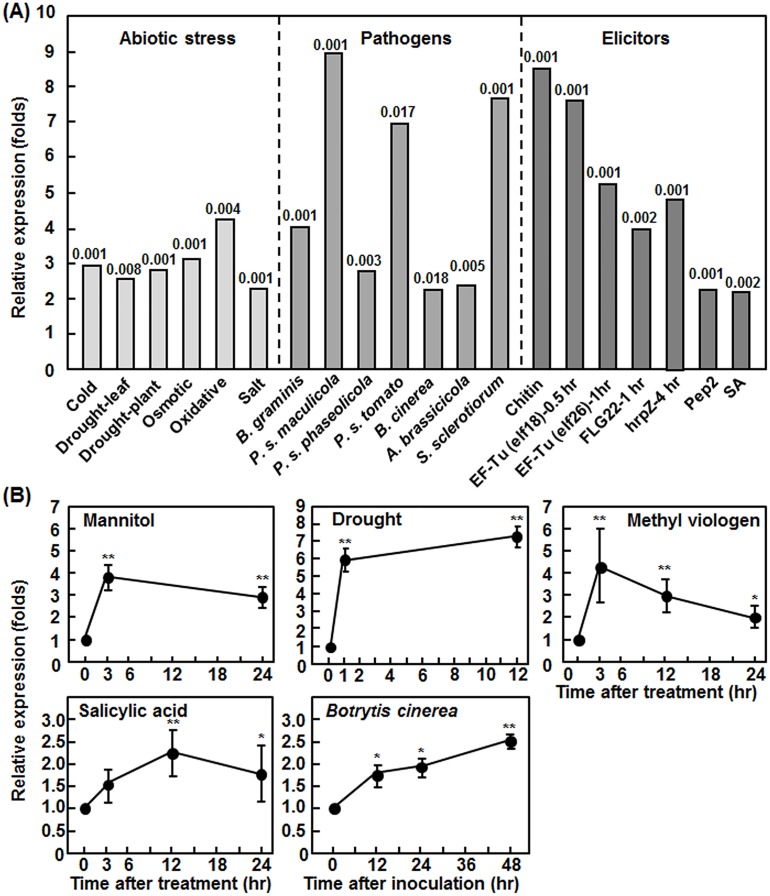
In our bioinformatics analyses of expression profiles of putative stress-responsive MAPKKKs, we found that a Raf-like MAPK kinase kinase gene, designated as Raf43, was highly induced by abiotic and biotic stresses. To analyze in detail the expression profile of Raf43 under stress conditions, we extracted the expression data of Raf43 (At3g46930) from public microarray databases using GENEVESTIGATOR platfrom. In our analyses, only expression changes of Raf43 in stress/elicitor-treated or pathogen-infected wild type Col-0 plants (except the Pst DC3000 experiment which used wild type Ws-0 plants) >2 folds with p values <0.05 were used to generate the expression profile of Raf43 in response to abiotic stress, pathogen infection and elicitor treatments (Fig 1A). Under abiotic stress conditions such as cold, drought, oxidative, osmotic and salt treatments, the expression levels of Raf43 in Col-0 plants were significantly increased by 2.26~4.23 folds over those in the controls (Fig 1A). In response to infection by different types of pathogens, e.g. biotrophic/hemibiotrophic (e.g. B. graminis and P. syringae) vs. necrotrophic (e.g. B. cinerea, A. brassicicola and S. sclerotiorum) and fungal (e.g. B. graminis, B. cinerea, A. brassicicola and S. sclerotiorum) vs. bacterial (e. g. P. syringae pvs. maculicola, phaseolicola and tomato) pathogens, the expression of Raf43 was significanlty induced in Col-0 and Ws-0 (only for P. syringae pv. tomato DC3000) plants, leading to 2.26~8.91 folds of increase over those in mock-inoculated plants (Fig 1A). In particular, relatively high levels of induction of Raf43 expression was observed after infections by bacterial pathogens P. syringae pv. maculicola and P. syringae pv. tomato DC3000 and necrotrophic fungal pathogen S. sclerotiorum, showing 8.91, 6.95 and 7.64 folds of increase, respectively (Fig 1A). Similarly, several well-known pathogen-associated molecular patterns such as fungal chitin and bacterial EF-Tu (elf18 and elf26) and FLG22, bacterial effectors such as HrpZ, Arabidopsis Pep2 protein and well-known defense signaling hormone SA, which are capable of activating both innate and inducible immune responses in Arabidopsis, induced significantly the expression of Raf43 in Col-0 plants, giving 2.16~8.48 folds of increase over those in the untreated plants (Fig 1A). Notably, the induced expression of Raf43 occurred rapidly in response to elicitor treatment; for example, significant induction of Raf43 expression was detected within one hour after treatment with EF-Tu and FLG22 (Fig 1A). To further confirm the expression profile of Raf43 in response to stress, we analyzed by qRT-PCR the expression patterns of Raf43 in Col-0 plants after treatment with mannitol, drought, MV and SA and infection with B. cinerea. The expression levels of Raf43 in mannitol- and MV-treated plants peaked with approximately 3 folds of increase over those in untreated plants at 3 hr and then decreased, whereas the expression level of Raf43 in drought stress-treated plants markedly increased with 6 folds over that in untreated control plants at 1 hr and continuously increased during a period of 12 hr (Fig 1B). When treated with 1 mM SA, the expression level of Raf43 increased at 3 hr, peaked with 2.35 folds of increase over that in untreated plants at 12 hr and then decreased (Fig 1B). Upon infection by B. cinerea, the expression level of Raf43 continuously increased during a period of 48 hr after inoculation (Fig 1B). These results confirmed the inducible expression feature of Raf43 revealed from analyses of public microarray data. It is worthy to note that the expression levels of Raf43 induced by SA treatment and by infection of B. cinerea in the qRT-PCR experiments were comparable to the levels observed from analyses of the public microarray data (Fig 1A and 1B). Taken together, these data indicate that Raf43 is responsive to multiple abiotic and biotic stresses and this stress-inducbile expression feature imply the involvement of Raf43 in abiotic and biotic stress response.
Fig 1. Responsiveness of Raf43 to biotic and abiotic stresses.
(A) Genevestigator-based analysis of expression profile of Raf43 in response to abiotic stress, pathogen infection and elicitor treatment. Expression data were extracted from Genevestigator microarray datasets obtained from experiments with ecotype Col-0 plants except for the P. syringae pv. tomato experiment, which was conducted with Ws-0 plants. The p values for the expression data between the treated/inoculated and control plants are shown above the columns. B. graminis, Blumeria graminis; P. s. maculicola, Pseudomonas syringae pv. maculicola; P. s. phaseolicola, P. syringae pv. phaseolicola; P. s. tomato, P. syringae pv. tomato DC3000; B. cinerea, Botrytis cinerea; A. brasicicola, Alternaria brassicicola; S. sclerotiorum, Sclerotinia sclerotiorum. (B) qRT-PCR-based analysis of expression patterns of Raf43 in response to treatments with mannitol, drought, methyl viologen or salicylic acid and to infection by B. cinerea. The expression level of Ubiquitin was used to normalize the Raf43 expression data and the relative expression was calculated as folds of expression in treated/inoculated plants vs. those in the control plants. Experiments were repeated with three independent biological samples at each time point and data represented are the means ± standard errors from three independent experiments. * and ** indicate the significant difference at p = 0.05 and p = 0.01, respectively, as compared with those in the controls.
Characterization of raf43-1 mutant and Raf43-overexpressing transgenic lines
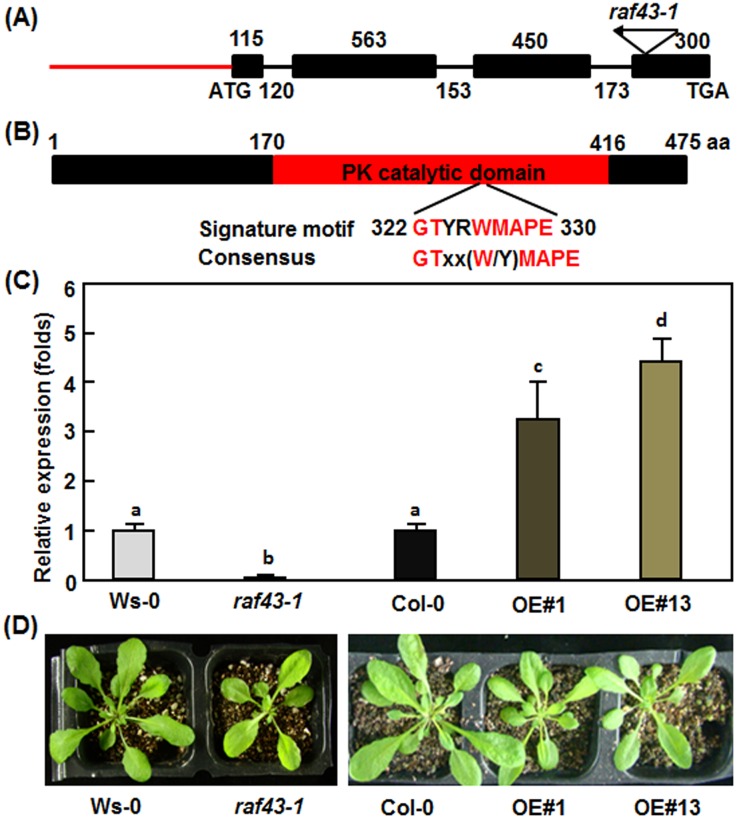
Raf43 is encoded by At3g46930 and the predicted ORF of Raf43 contains 4 exons and 3 introns (Fig 2A). Our cloning and sequencing confirmed the sequence of the predicted ORF of Raf43. The Raf43 gene encodes a 475 aa protein, which contains a typical protein kinase catalytic domain (170–416 aa) and a signature motif (322–330 aa) commonly present in Raf-like MAPKKKs [10] (Fig 2B). The protein structure feature indicates that Raf43 is a Raf-like MAPKKK and belongs to Group C of plants MAPKKKs [8]. To assess the biological function of Raf43, one T-DNA insertion line FLAG_505A06 in ecotype Ws-0 background [48] was analyzed. The T-DNA was inversely inserted in the last exon of the Raf43 gene (Fig 2A). PCR-based genotyping using a pair of gene-specific primers and a T-DNA primer identified individual plants that were homozygous at the T-DNA insertion site in the FLAG_505A06 line and this homozygous T-DNA insertion line was designated as raf43-1. qRT-PCR analysis revealed that the transcript level of Raf43 in raf43-1 plants was significantly reduced as compared with its wild type Ws-0 plants (Fig 2C), indicating that raf43-1 is a null mutant for Raf43. Meanwhile, we cloned the Raf43 ORF and inserted into pCAMBIA-991 vector under the control of CaMV 35S promoter to make a Raf43-overexpressing (OE) construct. The Raf43-OE construct was introduced into wild type Col-0 plants through floral dip transformation procedure and Raf43-overexpressing transgenic lines were obtained. Homozygous T3 lines with a single copy of the transgene were screened based on a 3:1 Hgr-resistant/Hgr-sensitive segregation ratio on selective medium and two independent lines, named Raf43-overexpressing OE#1 and Raf43-overexpressing OE#13 (thereafter referred as to Raf43-OE#1 and Raf43-OE#13), were chosen for further studies. qRT-PCR analysis showed that the transcript levels of Raf43 in Raf43-OE#1 and Raf43-OE#13 plants were significantly increased, leading to 3–4 folds of increase as compared with wild type Col-0 (Fig 2C). During our experiments, no any defect in growth and development was observed for the raf43-1 mutant plants and for the Raf43-OE#1 and Raf43-OE#13 plants when compared with their corresponding wild type Ws-0 and Col-0 plants, respectively (Fig 2D).
Fig 2. Structures of the Raf43 gene/protein and characterization of the raf43-1 mutant and the Raf43-overexpressing lines.
(A) Diagram showing the structure of Raf43 and the T-DNA location in raf43-1 mutant. Black boxes, black lines and red line indicate the exons, introns and putative promoter region, respectively. Sizes (bp) of the exons and introns are indicated above and below the diagram, respectively. The location and orientation of the T-DNA inserted in raf43-1 mutant line are shown. The start (ATG) and stop (TGA) codons are also indicated. (B) Diagram showing the structure of the Raf43 protein. The red box indicates the conserved protein kinase (PK) catalytic domain in Raf43 protein. Signature motif and its position in Raf43 protein and consensus of the signature motif commonly present in Raf-like MAPKKKs are shown. The amino acid positions are also indicated above the diagram. (C) Levels of the Raf43 transcripts in raf43-1 mutant and Raf43-overxpressing lines Raf43-OE#1 and Raf43-OE#13 plants. The transcript levels of Raf43 in four-week-old plants grown under normal conditions were analyzed by qRT-PCR using Raf43-specific primers and data were normalized by the transcript level of Ubiquitin. The transcript levels of Raf43 in wild type (Ws-0 and Col-0) plants were set as 1 and the levels in raf43 mutant and Raf43-overxpressing are shown as folds of the levels in wild type plants. Data represented are the means ± standard errors from three independent experiments. Different letters above the columns indicate significant difference at p = 0.05 between Ws-0 and raf43-1 plants and between Col-0 and Raf43-overexpressing plants. (D) Morphological and growth phenotypes of four-week-old plants of wild type (Ws-0 and Col-0), raf43-1 mutant and Raf43-overexpressing lines OE#1 and OE#13.
Raf43 is required for osmotic stress tolerance
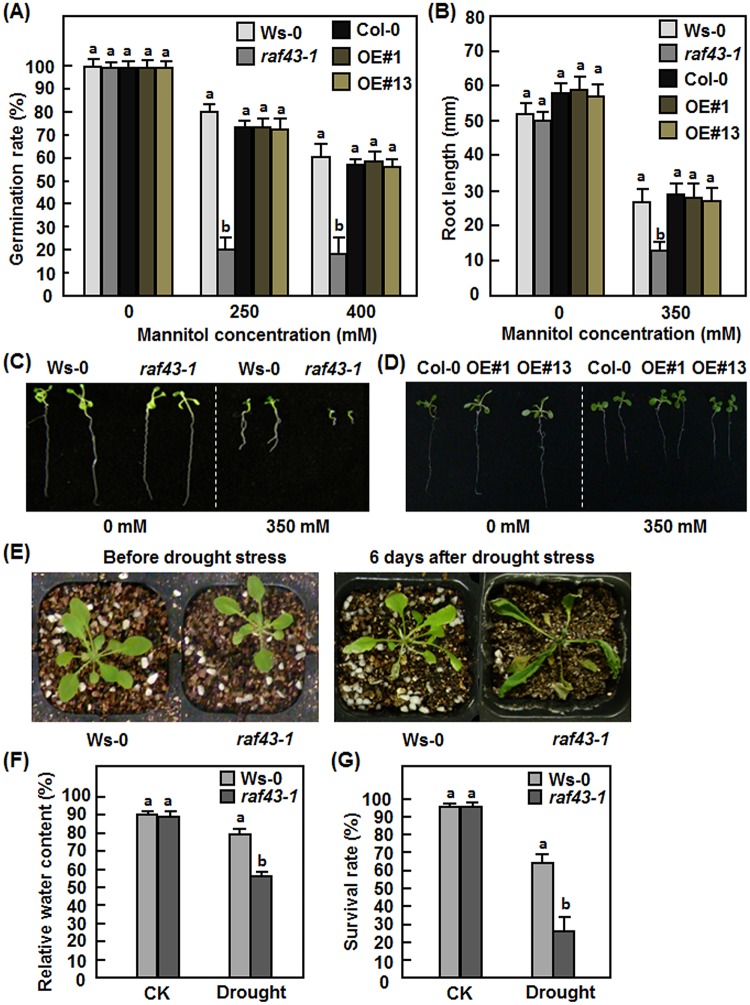
To explore the possible role of Raf43 in abiotic stress responses, we first examined and compared the seed germination and seedling root growth of the raf43-1 mutant line and the Raf43-OE#1 and Raf43-OE#13 lines with their corresponding wild types Ws-0 and Col-0 under osmotic stress condition mimicked by mannitol treatment. Germination rates of seeds from the raf43-1 mutant line, the Raf43-OE#1 and Raf43-OE#13 lines and their corresponding wild type Ws-0 and Col-0 was similar on mannitol-free 1/2MS medium but the germination rates of the raf43-1, Raf43-OE#1, Raf43-OE#13 and wild type seeds significantly decreased on 1/2MS medium supplemented with 200 mM or 400 mM mannitol as compared with those on mannitol-free medium (Fig 3A). In contrast, germination rates of the raf43-1 mutant seeds markedly decreased on 1/2MS medium supplemented with 200 mM or 400 mM mannitol, resulting in 70–75% of reduction as compared with the Ws-0 seeds (Fig 3A). No significant difference in germination rates of seeds from the Raf43-OE#1 and Raf43-OE#13 lines and wild type Col-0 was observed (Fig 3A). Similarly, after germination, root length of seedlings from the raf43-1 mutant line, the Raf43-OE#1 and Raf43-OE#13 lines and their corresponding wild types Ws-0 and Col-0 was similar when grown on mannitol-free 1/2MS medium but the root growth of the raf43-1, Raf43-OE#1, Raf43-OE#13 and wild type seedlings was significantly inhibited when grown on 1/2MS medium supplemented with 350 mM mannitol as compared with those on mannitol-free medium (Fig 3B–3D). Root growth of the raf43-1 seedlings was significantly inhibited on 1/2MS medium supplemented with 350 mM mannitol, leading to a 55% reduction in root length as compared with the Ws-0 seedlings (Fig 3B and 3C). No significant difference in root length of the Raf43-OE#1 and Raf43-OE#13 seedlings and the Col-0 seedlings was observed (Fig 3B and 3D). These observations indicate that knockout of Raf43 attenuated the osmotic stress tolerance but overexpression of Raf43 did not affect the osmotic stress tolerance, demonstrating the requirement of Raf43 in tolerance to osmotic stress in Arabidopsis.
Fig 3. Raf43 is required for osmotic and drought stress tolerance.
(A)–(D) Seed germination and root growth of seedlings grown on 1/2 MS supplemented with different concentrations of mannitol. (A) Seed germination rates on 1/2 MS supplemented with 0, 250 and 400 mM of mannitol. Seed germination was recorded when the radicles emerged completely from seed coat. (B) Root length of seedlings grown on 1/2 MS supplemented with 0 and 350 mM of mannitol. (C) and (D) Growth phenotype of seedlings grown on 1/2 MS supplemented with 0 and 350 mM mannitol. (E)-(G) Phenotype, relative water content and survival rates of soil-grown plants after drought stress treatment. (E) Phenotype of the wild type Ws-0 and mutant raf43-1 plants under drought stress condition. Three-week-old soil-grown plants were withheld from watering for drought stress treatment and allowed to grow for two weeks. Left photo shows the growth status of the plants before drought stress treatment while right photo shows phenotype of the plants at 7 days after drought stress treatment. (F) Relative water contents in leaf tissues of wild type Ws-0 and mutant raf43-1 plants grown under normal and drought stress condition at 7 days after treatment. (G) Survival rates of wild type Ws-0 and mutant raf43-1 plants grown under normal and drought stress condition at 14 days after treatment. Experiments were repeated three times and data presented in (A), (B), (F) and (G) are the means ± standard errors from three independent experiments. Different letters above the columns indicate significant difference at p = 0.05 between Ws-0 and raf43-1 plants and between Col-0 and Raf43-overexpressing plants.
Raf43 is required for drought stress tolerance
We next examined and compared the phenotype of soil-grown three-week-old raf43-1 mutant and its wild type Ws-0 plants in response to drought stress induced by withholding water. Before drought stress treatment, the growth status of the raf43-1 and wild type Ws-0 plants was similar (Fig 3E, left); however, most of the leaves of the raf43-1 plants were rolled, whereas only some old leaves of the wild type Ws-0 plants were slightly rolled at 7 days after withholding water (Fig 3E, right). Similar RWC in leaves of normally watered raf43-1 and Ws-0 plants was observed but withholding water for 7 days significantly reduced RWC in leaves of both the raf43-1 and Ws-0 plants (Fig 3F). However, RWC in leaves of the raf43-1 plants (56%) was significantly lower than that in the wild type Ws-0 plants (78%), leading to a reduction of 28% in RWC (Fig 3F). After 14-day withholding water, the survival rate of the raf43-1 plants (24%) was markedly decreased as compared with that of the wild type Ws-0 plants (63%), showing a reduction of 62% (Fig 3G). The normally watered raf43-1 and Ws-0 plants grown in the same chamber grew well with >95% of survival rate during the 14-day experimental period (Fig 3G). These data indicate that knockout of Raf43 resulted in reduced tolerance to drought stress, demonstrating that Raf43 is required for drought stress tolerance in Arabidopsis.
Raf43 is required for salt stress tolerance
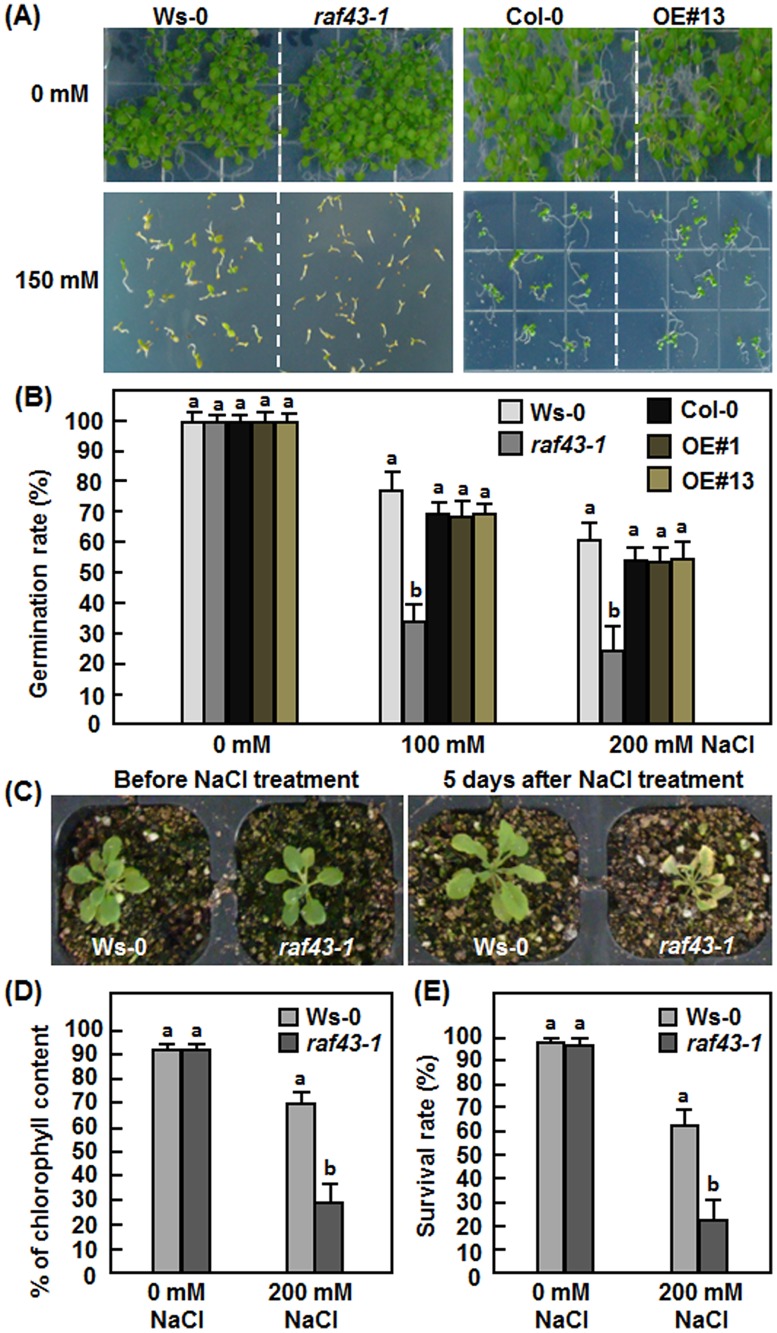
Possible function of Raf43 in salt stress tolerance was also evaluated by comparing the seed germination and root growth phenotype of the raf43-1 mutant line and the Raf43-OE#1 and Raf43-OE#13 lines with their corresponding wild type Ws-0 and Col-0 after treatment with NaCl. Germination rates of seeds from the raf43-1 mutant line, the Raf43-OE#1 and Raf43-OE#13 lines and their corresponding wild type Ws-0 and Col-0 was similar on NaCl-free 1/2MS medium but the germination rates of the raf43-1, Raf43-OE#1, Raf43-OE#13 and wild type seeds significantly decreased on 1/2MS medium supplemented with 100 mM or 200 mM NaCl as compared with those on mannitol-free medium (Fig 4A and 4B). In contrast, germination rates of the raf43-1 mutant seeds markedly decreased on 1/2MS medium supplemented with 100 mM or 200 mM NaCl, resulting in 60% of reduction as compared with the Ws-0 seeds (Fig 4A). No significant difference in germination rates of seeds from the Raf43-OE#1 and Raf43-OE#13 lines and wild type Col-0 was observed (Fig 4A and 4B). We further compared the phenotype of three-week-old soil-grown raf43-1 mutant and wild type Ws-0 plants in response to salt stress applied by drenching 200 mM NaCl. Before NaCl treatment, the growth status of the raf43-1 and wild type Ws-0 plants was similar (Fig 4C, left); however, most of the leaves of the raf43-1 plants showed yellowish necrotic symptoms, whereas only some leaves of the wild type Ws-0 plants were slightly yellowish at 5 days after NaCl treatment (Fig 4C, right). NaCl-induced damage in leaves of the raf43-1 and wild type Ws-0 plants resulted in significant reduction of the chlorophyll contents as compared with those in untreated control plants (Fig 4D). The reduction of chlorophyll contents in the NaCl-treated leaves of the raf43-1 plants was much higher than that in the wild type Ws-0 plants (Fig 4D). Chlorophyll content in leaves of NaCl-treated raf43-1 plants decreased to 32% of that in leaves without NaCl treatment, whereas chlorophyll content in NaCl-treated leaves of the wild type Ws-0 was about 76% of that in leaves without NaCl treatment (Fig 4D). At 10 days after NaCl treatment, the survival rate of the raf43-1 plants (21%) was markedly decreased as compared with that of the wild type Ws-0 plants (61%), showing a reduction of 66% (Fig 4E). No difference was observed in survival rates of the raf43-1 and Ws-0 plants (>95%) without NaCl treatment during the 7-day experimental period (Fig 4E). These observations indicate that knockout of Raf43 attenuated the salt stress tolerance whereas overexpression of Raf43 had no effect, suggesting that Raf43 is also required for salt stress tolerance in Arabidopsis.
Fig 4. Raf43 is required for salt stress tolerance.
(A) and (B) Seed germination on 1/2 MS supplemented with different concentrations of NaCl. (A) Seed germination and seedling growth on 1/2 MS supplemented with 0 and 150 mM of NaCl. Photos were taken at 10 days after germination. (B) Germination rates of seeds on 1/2 MS supplemented with 0, 100 and 200 mM of NaCl. Seed germination was recorded when the radicles emerged completely from seed coat. (C)-(E) Phenotype, chlorophyll content and survival rates of soil-grown plants after NaCl treatment. (C) Phenotype of the wild type Ws-0 and mutant raf43-1 plants under drought stress condition. Three-week-old soil-grown plants were drenched with 0 or 200 mM NaCl and allowed to grow for 10 days. Left photo shows the growth status of the plants before NaCl treatment while right photo shows phenotype of the plants at 5 days after NaCl treatment. (D) Percentages of chlorophyll content in leaf tissues of wild type Ws-0 and mutant raf43-1 plants grown under normal and NaCl stress condition at 5 days after treatment. (E) Survival rates of wild type Ws-0 and mutant raf43-1 plants grown under normal and drought stress condition at 10 days after treatment. Experiments were repeated three times and data presented in (B), (D) and (E) are the means ± standard errors from three independent experiments. Different letters above the columns indicate significant difference at p = 0.05 between Ws-0 and raf43-1 plants and between Col-0 and Raf43-overexpressing plants.
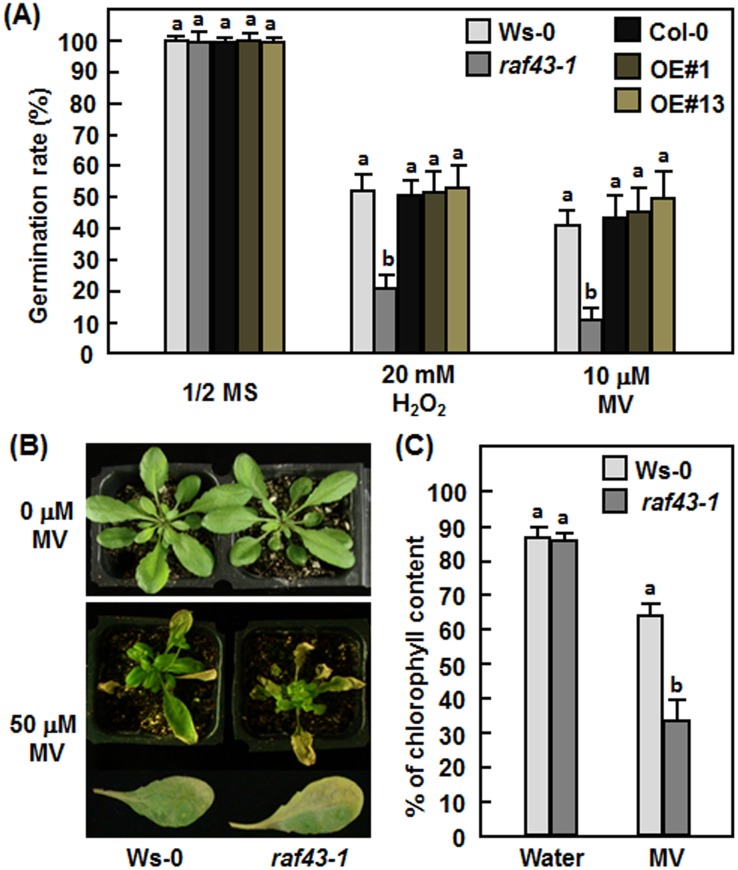
Raf43 is required for oxidative stress tolerance
Because expression of Raf43 was induced by oxidative stress (Fig 1), we thus examined whether mutation or overexpression of Raf43 would affect tolerance to oxidative stress during seed germination or in mature Arabidopsis leaves. In 1/2MS medium without supplementation of MV or H2O2, seeds of the raf43-1 mutant and the Raf43-OE#1 and Raf43-OE#13 lines showed similar germination rates to those of the corresponding wild type Ws-0 and Col-0 seeds (Fig 5A). In 1/2MS medium supplemented with 20 mM H2O2 or 10 μM MV, germination rates of seeds from the raf43-1 mutant line, the Raf43-OE#1 and Raf43-OE#13 lines and the wild types Ws-0 and Col-0 lines were decreased significantly, but seeds of the raf43-1 mutant line showed a lower germination rate than the wild type Ws-0 seeds (Fig 5A). At 6 days, only 20% and 10% of the raf43-1 mutant seeds had germinated in the presence of 20 mM H2O2 or 10 mM MV, respectively, while 40–50% of wild type Ws-0 seeds had germinated (Fig 5A). By contrast, germination rates for seeds of the Raf43-OE#1 and Raf43-OE#13 lines in the presence of 20 mM H2O2 or 10 μM MV were comparable to those of the wild type Col-0 seeds (Fig 5A). We further assessed the oxidative stress tolerance in mature leaves of the raf43-1 and wild type Ws-0 plants to MV. Without treatment with MV, both the raf43-1 and wild type Ws-0 plants grew well; however, exogenous application of 50 μM MV by foliar spraying caused significant necrosis or bleaching, indicative of oxidative damage, on leaves of soil-grown four-week-old raf43-1 and Ws-0 plants (Fig 5B). Oxidative damage caused by exogenously applied MV on leaves of the raf43-1 plants was much more pronounced than on leaves of the wild type Ws-0 plants (Fig 5B). MV-induced oxidative damage in leaves of the raf43-1 and wild type Ws-0 plants resulted in significant reduction of the chlorophyll contents as compared with those in untreated control plants (Fig 5C). The reduction of chlorophyll contents in the MV-treated leaves of the raf43-1 plants was much evident than that in the wild type Ws-0 plants (Fig 5C). Chlorophyll content in MV-treated leaves of the raf43-1 plants decreased to 39% of that in leaves without MV treatment, whereas chlorophyll content in MV-treated leaves of the wild type Ws-0 was about 71% of that in leaves without MV treatment (Fig 5C). These results indicate that knockout of Raf43 could weaken oxidative stress tolerance, leading to an increased level of cellular damage under oxidative stress conditions but overexpression of Raf43 did not affect the oxidative stress tolerance in Arabidopsis.
Fig 5. Raf43 is required for oxidative stress tolerance.
(A) Germination rates of seeds on 1/2 MS supplemented with 20 mM H2O2 or 10 μM methyl viologen (MV). Seed germination was recorded when the radicles emerged completely from seed coat. (B) and (C) Phenotype and chlorophyll contents in leaf tissues of the wild type Ws-0 and mutant raf43-1 plants after treatment with 50 μM MV. Representative leaves showing typical yellowish and necrotic symptom from MV-treated Ws-0 and raf43-1 plants were also shown. Three-week-old soil-grown plants were treated by foliar spraying with 50 μM MV or water as a control and allowed to grow for 7 days. Photos were taken at 5 days after treatment and leaf samples were collected for analyzing chlorophyll contents. Experiments were repeated three times and data presented in (A) and (C) are the means ± standard errors from three independent experiments. Different letters above the columns indicate significant difference at p = 0.05 between Ws-0 and raf43-1 plants and between Col-0 and Raf43-overexpressing plants.
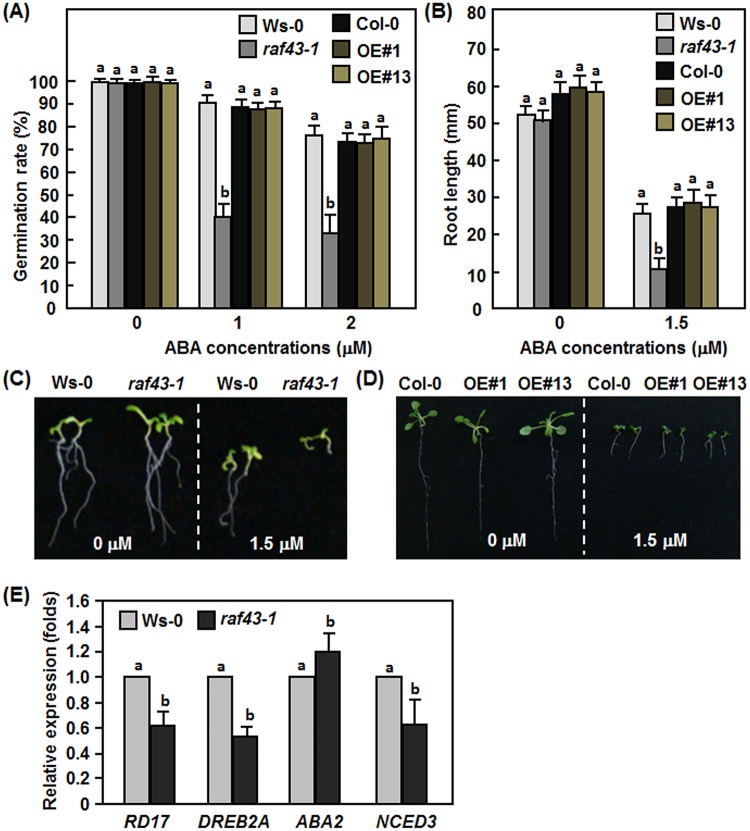
Raf43 is involved in ABA response
Considering that Raf43 is required for multiple abiotic stress tolerance as mentioned above and that ABA is an important stress hormone involved in regulating abiotic stress response [49], we therefore examined the possible involvement of Raf43 in ABA response. For this purpose, seed germination and root growth of the raf43-1 mutant line and the Raf43-OE#1 and Raf43-OE#13 lines in response to exogenous ABA were assayed and compared with their corresponding wild types Ws-0 and Col-0. Germination rates of seeds from the raf43-1 line, the Raf43-OE#1 and Raf43-OE#13 lines and their corresponding wild type Ws-0 and Col-0 were similar when germinated on ABA-free 1/2MS medium but the germination rates decreased significantly on 1/2MS medium supplemented with 1 μM or 2 μM ABA as compared with those on ABA-free medium (Fig 6A). However, germination rates of the raf43-1 mutant seeds markedly decreased on 1/2MS medium supplemented with 1 μM or 2 μM ABA, resulting in 55–60% of reduction as compared with the Ws-0 seeds (Fig 6A). No significant difference in germination rates of seeds from the Raf43-OE#1 and Raf43-OE#13 lines and wild type Col-0 was observed in the presence of ABA (Fig 6A). After germination, root length of the raf43-1 seedlings, the Raf43-OE#1 and Raf43-OE#13 seedlings and their corresponding wild type Ws-0 and Col-0 seedling was similar when grown on ABA-free 1/2MS medium but the root growth was significantly inhibited when grown on 1/2MS medium supplemented with 1.5 μM ABA as compared with those on ABA-free medium (Fig 6B–6D). Root growth of the raf43-1 seedlings was significantly inhibited on 1/2MS medium supplemented with 1.5 μM ABA, leading to a 56% of reduction as compared with the Ws-0 seedlings (Fig 6B and 6C). No significant difference in root length of the Raf43-OE#1 and Raf43-OE#13 seedlings and the wild type Col-0 seedlings was observed (Fig 6B and 6D). These observations indicate that knockout of Raf43 resulted in increased ABA sensitivity but overexpression of Raf43 had no effect on ABA response in Arabidopsis.
Fig 6. Raf43 is involved in ABA response.
(A) Seed germination rates on 1/2 MS supplemented with 0, 1 and 2 μM of ABA. Seed germination was recorded when the radicles emerged completely from seed coat. (B) Root length of seedlings grown on 1/2 MS supplemented with 0 and 1.5 μM of ABA. (C) and (D) Root phenotype of seedlings grown on 1/2 MS supplemented with 0 and 1.5 μM of ABA. (E) Expression of stress-responsive and ABA biosynthesis-related genes in WS-0 and raf43-1 plants. Leaf samples were collected form four-week-old soil-grown plants for analyzing gene expression through qRT-PCR. Expression data were normalized by the transcript level of Ubiquitin. The expression levels of Raf43 in wild type Ws-0 plants was set as 1 and the levels in raf43 mutant plants are shown as folds of the levels in wild type plants. Experiments were repeated three times and data presented in (A), (B) and (E) are the means ± standard errors from three independent experiments. Different letters above the columns indicate significant difference at p = 0.05 between Ws-0 and raf43-1 plants and between Col-0 and Raf43-overexpressing plants.
To gain information on the possible mechanism of action of Raf43 in abiotic stress response, we analyzed through qRT-PCR and compared the expression of four selected stress-responsive and ABA biosynthesis-related genes such as NCED3 [50], ABA2 [51, 52], RD17 (COR47) [53], and DREB2A [54] in raf43-1 and Ws-0 plants grown under normal condition. As shown in Fig 6E, the expression levels of the stress-responsive RD17 and DREB2A in raf43-1 plants were significantly reduced, leading to 50% and 40% of reduction, respectively, as compared with those in Ws-0 plants (Fig 6E). The ABA biosynthetic genes NCED3 and ABA2 showed different expression patterns in the raf43-1 plants. The expression level of NCED3 in the raf43-1 plants reduced markedly, leading to a 39% of reduction whereas the expression level of ABA2 showed a slight increase (19%), as compared with those in Ws-0 plants (Fig 6E). These results indicate that knockout of Raf43 resulted in altered expression of stress-responsive and ABA biosynthesis-related genes.
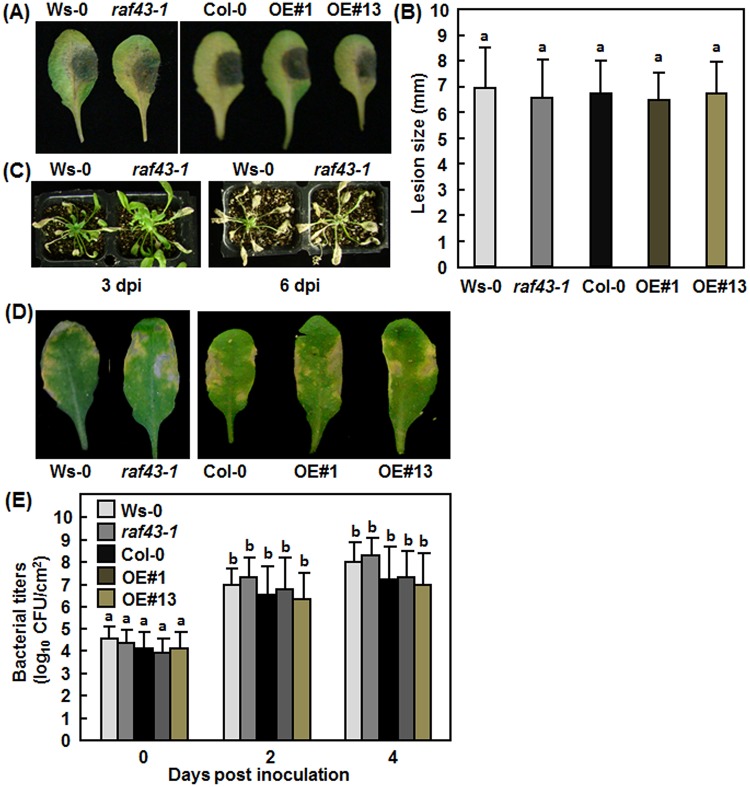
Raf43 is not required for resistance against B. cinerea and P. syringae pv. tomato DC3000
The induction of Raf43 by pathogen infection and elicitor treatment (Fig 1) led us to examine whether Raf43 plays a role in resistance against different pathogens. Detached leaf inoculation and whole plant inoculation assays were performed to compare the disease phenotype of the raf43-1 mutant plants and the Raf43-OE#1 and Raf43-OE#13 plants with their corresponding wild type plants after inoculation with B. cinerea. In the detached leaf inoculation assays, no significant difference in size of necrotic lesions caused by B. cinerea was observed between Ws-0 and raf43-1 plants and between Col-0 and Raf43-OE#1 and Raf43-OE#13 plants (Fig 7A and 7B). Similar results were also obtained from the whole plant inoculation assays. Typical yellowish necrotic lesions were observed on leaves of wild type Ws-0 and mutant raf43-1 plants at 3 days and these diseased plants started to die at 6 days after inoculation by foliar spraying of B. cinerea spores suspension (Fig 7C). On the other hand, we also explored the possible function of Raf43 in resistance against a bacterial pathogen by comparing disease phenotypes of the raf43-1 mutant plants and the Raf43-OE#1 and Raf43-OE#13 plants with their corresponding wild types Ws-0 and Col-0 plants after inoculation with a normally virulent strain of P. syringae pv. tomato DC3000. In these assays, typical chlorotic lesions were observed in inoculated leaves of the Ws-0 and raf43-1 plants and the Col-0 and Raf43-E#1/Raf43-OE#13 plants at 4 days after inoculation (Fig 7D). Measurement of bacterial titers in inoculated leaves showed no significant difference in bacterial proliferation between the Ws-0 and raf43-1 plants and the Col-0 and Raf43-OE#1/Raf43-OE#13 plants at 2 and 4 days after inoculation (Fig 7E). These results indicate that knockout or overexpression of Raf43 did not affect the response of Arabidopsis plants to infection of B. cinerea and P. syringae pv. tomato DC3000, implying that Raf43 is not required for resistance to these two pathogens.
Fig 7. Raf43 is not required for resistance to B. cinerea and P. syringae pv. tomato DC3000.
(A)-(C) Phenotype of disease caused by B. cinerea. (A) Disease phenotype on leaves of different genotype plants in detached leaf inoculation assays. (B) Lesion sizes on leaves from (A). Leaves were detached from four-week-old plants and inoculated by dropping 5 μL of B. cinerea spore suspension (1×105 spores/mL). Photos were taken and lesion sizes were recorded at 5 days after inoculation. (C) Disease phenotype on wild type Ws-0 and mutant raf43 plants. Four-week-old plants were inoculated by foliar spraying with B. cinerea spore suspension (1×105 spores/mL) and photos were taken at 3 and 6 days after inoculation. dpi, days post inoculation. (D) and (E) Phenotype of disease caused by P. syringae pv. tomato DC3000. (A) Representative disease symptom on leaves of different genotype plants. (B) Bacterial titers in inoculated leaves. Four-week-old plants were inoculated by infiltration with P. syringae pv. tomato DC3000 (OD600 = 0.002). Photos were taken 4 days after inoculation. Leaf samples were collected at 0, 2, and 4 days after inoculation and bacterial growth was measured. Experiments were repeated three times and the data presented in (B) and (E) are the means ± standard errors from three independent experiments. Different letters above the columns indicate significant difference at p = 0.05 between Ws-0 and raf43-1 plants and between Col-0 and Raf43-overexpressing plants.
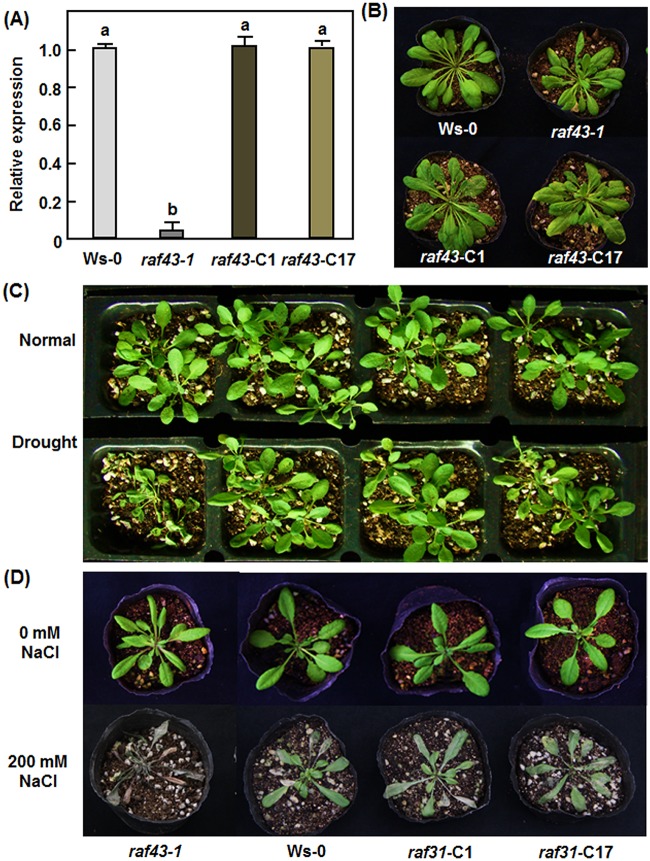
Native promoter-driven expression of Raf43 restores the stress tolerance in raf43 plants
Because only one T-DNA insertion line was used for analysis of the Raf43 function, genetic complementation was thus carried out to determine whether the decreased abiotic stress tolerance that we observed in raf43 plants is indeed the result of the mutation within the Raf43 gene. A fusion of 1295 bp native promoter region and the Raf43 ORF was introduced into raf43-1 mutant background. Homozygous T3 lines with a single copy of the transgene were screened based on a 3:1 Hgr-resistant/Hgr-sensitive segregation ratio and two independent complementation lines, named Raf43-C1 and Raf43-C17, were chosen for further studies. qRT-PCR analysis showed that the transcript levels of Raf43 in Raf43-C1 and Raf43-C17 plants were comparable to that in Ws-0 plants but significantly higher than that in raf43-1 plants (Fig 8A). The growth and morphology of the Raf43-C1 and Raf43-C17 plants were undistinguishable from the Ws-0 and raf43-1 plants (Fig 8B). In drought and salt stress assays, the Raf43-C1 and Raf43-C17 plants displayed similar phenotypes while the raf43-1 plants showed significant stress damages, as compared with the Ws-0 plants (Fig 8C and 8D). These data indicate that the native promoter-driven expression of Raf43 fully complemented the phenotypes observed in the raf43-1 plants, demonstrating that the decreased abiotic stress tolerance in raf43 plants is indeed caused by the mutation within the Raf43 gene.
Fig 8. Native promoter-driven expression of Raf43 complemented the phenotype of raf43-1 plants under stress conditions.
(A) Levels of the Raf43 transcripts in Raf43-complementation lines Raf43-C1 and Raf43-C17 plants. The transcript levels of Raf43 in four-week-old plants grown under normal conditions were analyzed by qRT-PCR using Raf43-specific primers and data were normalized by the transcript level of Ubiquitin. The transcript level of Raf43 in Ws-0 plants was set as 1 and the levels in raf43-1, Raf43-C1 and Raf43-C17 plants are shown as folds of the levels in Ws-0 plants. Data represented are the means ± standard errors from three independent experiments and different letters above the columns indicate significant difference at p = 0.05. (B) Morphological and growth phenotypes of four-week-old Ws-0, raf43-1, Raf43-C1 and Raf43-C17 plants. (C) Phenotype of the Ws-0, raf43-1, Raf43-C1 and Raf43-C17 plants under drought stress condition. Three-week-old soil-grown plants were withheld from watering for drought stress treatment and allowed to grow for 7 days. Photo shows the phenotype of the plants at 7 days after drought stress treatment. (D) Phenotype of the Ws-0, raf43-1, Raf43-C1 and Raf43-C17 plants under salt stress condition. Three-week-old soil-grown plants were drenched with 200 mM NaCl and allowed to grow for 7 days. The photo shows phenotype of the plants at 7 days after NaCl treatment. Repeated experiments for (C) and (D) showed similar results.
Discussion
The plant MAPKKKs have great sequence diversity and are categorized either as MEKKs, Raf and ZIK subfamilies or as three groups (Groups A, B and C) [8, 10]. However, only a few of Raf-like MAPKKKs in plants have been studied and most of the characterized Raf-like MAPKKKs belong to the Group B of Raf-like MAPKKKs, including Arabidopsis CTR1, EDR1, SIS8/AT6 and MAP3Kδ4 and rice ERD1 and DSM1 [32, 33, 39–42, 44, 55]. To date, the sole Group C Raf-like MAPKKK that has been functionally characterized in plants is the rice ILA1 [43]. In the present study, we identified an Arabidopsis stress-responsive Group C Raf-like MAPKKK gene, Raf43, which can be induced by multiple abiotic and biotic stresses. Functional analyses using a T-DNA insertion mutant raf43-1 demonstrated that Raf43 is required for tolerance to multiple abiotic stresses including drought, osmotic, salt and oxidative stresses. Our findings provide new insights into the biological roles of the functionally uncharacterized Group C Raf-like MAPKKKs in plants.
Our interesting on Raf43 came from bioinformatics analyses of publicly available microarray data to identify putative stress-responsive MAPKKK genes. Expression profiling analyses using publicly available microarray data and our qRT-PCR data clearly demonstrated that Raf43 is responsive to multiple abiotic stresses including drought, oxidative, osmotic and salt stress (Fig 1). Phenotype analyses of the raf43-1 mutant and two Raf43-overexpressing lines revealed that Raf43 is necessary but not sufficient for tolerance to drought, osmotic, oxidative and salt stresses and thus acts as a positive regulator of multiple abiotic stress responses. This is different from the function of SIS8/AT6, which negatively regulates salt tolerance in Arabidopsis [39]. Several lines of evidence presented in this study support the requirement of Raf43 for tolerance to abiotic stress response. Firstly, seed germination and seedling root growth of raf43-1 line was significantly inhibited on medium supplemented with mannitol, NaCl, H2O2 or MV (Figs 3, 4 and 5). Secondly, reduced tolerance to drought, salt and oxidative stress was observed in the soil-grown mature raf43-1 plants upon withholding of water, drenching of NaCl solution or foliar spraying of MV (Figs 3, 4 and 5). This is in agreement with the function of rice DSM1, which was found to positively regulate drought and oxidative stress tolerance [42]. The native promoter-driven expression of Raf43 fully complemented the phenotypes of the raf43-1 plants under abiotic stress conditions, demonstrating that the decreased abiotic stress tolerance in raf43-1 plants is indeed caused by the mutation within the Raf43 gene (Fig 8). Thirdly, the expression of RD17 and DREB2A, two stress-responsive genes [53, 54], was downregulated in raf43-1 plants (Fig 6E), indicating that Raf43 may be involved in stress-responsive signaling pathway. Fourthly, mutation in Raf43 resulted in increased ABA sensitivity as seed germination and seedling root growth of raf43-1 line was inhibited markedly in the presence of exogenous ABA (Fig 6). It is well known that, as a stress hormone, ABA plays a key role in the regulation of many physiological processes in response to abiotic stress response [49, 56]. The altered expression of NCED3 and ABA2, two stress-responsive genes [50–52], in raf43-1 plants may indicate the involvement of Raf43 in ABA response as well as in ABA-mediated signaling pathway. This differs from the function of SS8/AT6 that negatively regulates salt stress tolerance in an ABA-independent manner [39]. In addition, our functional analyses in this study also demonstrate that Raf43 has pleiotropic effects on tolerance to multiple abiotic stresses. This is similar to SIS7/AT6, which was shown to play roles in salt and high sugar response [39, 40], and to rice DSM1, which is required for both drought and oxidative stress [42].
Both microarray data- and qRT-PCR-based expression profiling analyses indicated that the expression of Raf43 was significantly induced by infection of some fungal and bacterial pathogens and treatment with elicitors including some well-known pathogen-associated molecular patterns and defense signaling hormone SA (Fig 1) [57], implying a role for Raf43 in disease resistance. Surprisingly, neither the raf43-1 plants nor the Raf43-overexpressing plants exhibited any difference in disease phenotypes to the wild type plants after inoculation with necrotrophic fungal pathogen B. cinerea and hemibiotrophic bacterial pathogen P. syringae pv. tomato DC3000 (Fig 7). It is thus unlikely that Raf43 plays a role in resistance against these two pathogens. However, the involvement of Raf43 in biotic stress response cannot be ruled out at present and needs to be investigated further using more pathogens with different infection styles. Furthermore, the significant induction of Raf43 by some of pathogen-associated molecular patterns such as FLG22 and EF-Tu may imply that Raf43 has a function in regulating innate immunity in Arabidopsis.
It was previously reported that overexpression of tobacco NPK1 and Arabidopsis MAP3Kδ4 in stable transgenic plants and transient expression of tomato MAPKKKε and N. benthamiana NbMAPKKKα, NbMAPKKKβ and NbMAPKKKγ in leaves of N. benthamiana resulted in increased tolerance to abiotic stresses and accelerated programmed cell death, respectively [28, 30, 55, 58, 59]. Similarly, overexpression of DSM1, encoding for a Group B Raf-like MAPKKK, in transgenic rice increased the tolerance to drought stress at the seedling stage [42]. However, we did not observe any alteration in abiotic and biotic stress response in the two Raf43-overexpressing lines. The reason for this phenomenon is unclear. The possibility that sequence errors of the transgene were introduced during the cloning and vector construction processes and thus resulted in misexpression of Raf43 in Raf43-overexpressing lines can be ruled out as no any error in the transgene was identified in these overexpression lines by re-sequencing. The most possibility to explain the phenomenon that overexpression of Raf43 did not result in any alteration in abiotic stress response should be associated with the biochemical nature of the Raf43 protein itself. Some of Raf-like MAPKKKs such as Arabidopsis CTR1 and rice ILA1 have been reported to possess kinase activities and have autophosphorylation activity in vitro [43, 60, 61]. It was also found recently that phosphorylation of Arabidopsis MEKK1 via Ca2+ signaling is critical to its function in cold stress response [62]). Furthermore, the kinase catalytic domains in Raf-like MAPKKKs including Arabidopsis CTR1 and EDR1 and rice DSM1 were shown to have kinase activity [42, 60, 61]. Raf43 contains a typical kinase catalytic domain at C-terminal (Fig 2B). It is thus likely that the activity of Raf43 as a predicted MAPKKK needs to be activated via phosphorylation by itself or upstream signals and thus simply increases of the transcript or protein through the overexpression way cannot exert its biochemical function in the signaling events. Similar phenomenon was also observed for some MAPKKs. For example, overexpression of MKK3 in Arabidopsis or transient expression of tomato SlMKK2 and SlMMK4 in N. benthamiana did not affect the disease phenotype of P. syringae pv. tomato DC3000 and the appearance of programmed cell death, respectively, whereas overexpression of constitutively active forms of these MAPKKs conferred clear phenotypes [63, 64]. Alternatively, the kinase activity of full-length Raf43 may be another factor that affects its kinase activity in the Raf43-overexpressing plants. It was found that the full-length Arabidopsis MEKK1 showed almost no activity but the kinase domain of MEKK1 had a strong ability to phosphorylate its substrate MKK2 [62]. Further studies on biochemical characterization of the kinase activity of Raf43 and phenotypical analyses of plants overexpressing the kinase domain of Raf43 will be helpful to address these questions.
As components acting at the top of MAPK cascades, MAPKKKs are generally believed to phosphorylate their downstream MAPKKs to relay the signal transduction events. In this regard, several members of the MEKK group have been shown to function as MAPKKKs upstream of MAPKKs in MAPK cascades. It has been demonstrated that the Arabidopsis MEKK1 and YODA and the tobacco NPK1, all belonging to the MEKK group of MAPKKKs, can form typical MAPK cascades, e.g. MEKK1–MKK4/MKK5–MPK3/MPK6 [65], YDA—MKK4/MKK5–MPK3/MPK6 [18, 66], and NPK1–NQK1–NRK1 [67]. However, traditional MAPK cascade has not been established yet for members of the Raf-like MAPKKKs. Instead, several studies have indicated that some of the identified Raf-like MAPKKKs do not function as MAPKKKs. For example, CTR1 was reported to inhibit MKK9–MPK3/MPK6 activation [36], whereas EDR1 was found to negatively regulate the MKK4/MKK5-MPK3/MPK6 cascade [38]. In our yeast two-hybrid-based pairwise interaction experiment, no significant interaction of Raf43 to any of 10 Arabidopsis MAPKKs was detected (data not shown). This is in agreement with the observation that the rice ILA1, a group C of Raf-like MAPKKK, did not interact with all rice MAPKKs [43]. Thus, it is likely that members of the Raf-like MAPKKKs may not follow the traditional MAPK cascades; instead, they exert their function by directly interacting with specific targets, which are not MAPKKs. This hypothesis is supported by the identification of the ILA1- and SIS8/AT6-interacting proteins. ILA1 was shown to interact with six closely related proteins with unknown function while SIS8/AT6 was found to interact with a UDP-glucosyltransferase UGT72E1 [40, 43]. Another, direct phosphorylation of a transcription factor by a MAPKKK has also been previously reported [68]. Therefore, it is possible that Raf43 may interact with downstream targets rather than MAPKKs to modulate the signaling events. Further screening and identification of Raf43-interacting proteins will provide new information on the mechanism for the function of Raf43 in abiotic stress response.
Acknowledgments
This work was supported by the National High-Tech R & D Program (No. 2012AA101505), the National Transgenic Major Project of China (No. 2011ZX08009-003-001), the National Natural Science Foundation (No. 31272028) and the Research Fund for the Doctoral Program of Higher Education of China (20120101110070).
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the National High-Tech R & D Program (No. 2012AA101505), the National Transgenic Major Project of China (No. 2011ZX08009-003-001), the National Natural Science Foundation (No. 31272028) and the Research Fund for the Doctoral Program of Higher Education of China (20120101110070).
References
- 1. Faulkner C, Robatzek S. Plants and pathogens: putting infection strategies and defence mechanisms on the map. Curr Opin Plant Biol. 2012; 15:699–707. 10.1016/j.pbi.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 2. Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot. 2013; 64:445–458. 10.1093/jxb/ers354 [DOI] [PubMed] [Google Scholar]
- 3. Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R. Abiotic and biotic stress combinations. New Phytol. 2014; 203:32–43. 10.1111/nph.12797 [DOI] [PubMed] [Google Scholar]
- 4. Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010; 61:621–649. 10.1146/annurev-arplant-042809-112252 [DOI] [PubMed] [Google Scholar]
- 5. Sinha AK, Jaggi M, Raghuram B, Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav. 2011; 6:196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rasmussen MW, Roux M, Petersen M, Mundy J. MAP kinase cascades in Arabidopsis innate immunity. Front Plant Sci. 2012; 3:169 10.3389/fpls.2012.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013; 51:245–266. 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- 8. MAPK group. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002; 7:301–308. [DOI] [PubMed] [Google Scholar]
- 9. Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006; 11:192–198. [DOI] [PubMed] [Google Scholar]
- 10. Jonak C, Okresz L, Bogre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002; 5:415–424. [DOI] [PubMed] [Google Scholar]
- 11. Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 2010; 17:139–153. 10.1093/dnares/dsq011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kong X, Lv W, Zhang D, Jiang S, Zhang S, Li D. Genome-wide identification and analysis of expression profiles of maize mitogen-activated protein kinase kinase kinase. PLoS One. 2013; 8: e57714 10.1371/journal.pone.0057714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin Z, Wang J, Wang D, Fan W, Wang S, Ye W. The MAPKKK gene family in Gossypium raimondii: genome-wide identification, classification and expression analysis. Int J Mol Sci. 2013; 14:18740–18757. 10.3390/ijms140918740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Y, Wang C, Yang B, Wu F, Hao X, Liang W, et al. Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J Exp Bot. 2014; 65:2171–88. 10.1093/jxb/eru092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu J, Wang J, Pan C, Guan X, Wang Y, Liu S, et al. Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS One. 2014; 9:e103032 10.1371/journal.pone.0103032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krysan PJ, Jester PJ, Gottwald JR, Sussman MR. An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell. 2002; 14:1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lukowitz W, Roeder A, Parmenter D, Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004; 116: 109–119. [DOI] [PubMed] [Google Scholar]
- 18. Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004; 304: 1494–1497. [DOI] [PubMed] [Google Scholar]
- 19. Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, et al. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004; 15:141–152. [DOI] [PubMed] [Google Scholar]
- 20. Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature dependent cell death in Arabidopsis. J Biol Chem. 2006; 281: 36969–36976. [DOI] [PubMed] [Google Scholar]
- 21. Nakagami H, Soukupova H, Schikora A, Zarsky V, Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem. 2006; 28: 38697–38704. [DOI] [PubMed] [Google Scholar]
- 22. Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007; 143:661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, et al. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008; 18:1190–1198. 10.1038/cr.2008.300 [DOI] [PubMed] [Google Scholar]
- 24. Kong Q, Qu N, Gao M, Zhang Z, Ding X, Yang F, et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell. 2012; 24:2225–2236. 10.1105/tpc.112.097253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin H, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang S, Staskawicz B, et al. NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev Cell. 2002; 3:291–297. [DOI] [PubMed] [Google Scholar]
- 26. del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004; 23: 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakagami H, Kiegerl S, Hirt H. OMTK1, a novel MAPKKK, channels oxidative stress signaling through direct MAPK interaction. J Biol Chem. 2004; 279:26959–26966. [DOI] [PubMed] [Google Scholar]
- 28. Melech-Bonfil S, Sessa G. Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 2010; 64: 379–391. [DOI] [PubMed] [Google Scholar]
- 29. Oh CS, Pedley KF, Martin GB. Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKKα. Plant Cell. 2010; 22: 260–272. 10.1105/tpc.109.070664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hashimoto M, Komatsu K, Maejima K, Okano Y, Shiraishi T, Ishikawa K, et al. Identification of three MAPKKKs forming a linear signaling pathway leading to programmed cell death in Nicotiana benthamiana . BMC Plant Biol. 2012; 12:103 10.1186/1471-2229-12-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. King SR, McLellan H, Boevink PC, Armstrong MR, Bukharova T, Sukarta O, et al. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell. 2014; 26:1345–1359. 10.1105/tpc.113.120055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993; 72:427–441. [DOI] [PubMed] [Google Scholar]
- 33. Frye CA, Tang D, Innes RW. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA. 2001; 98:373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998; 95:5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, et al. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem. 2003; 278:34725–34732. [DOI] [PubMed] [Google Scholar]
- 36. Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008; 451: 789–795. 10.1038/nature06543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gibson S, Laby R, Kim D. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1 . Biochem Biophys Res Commun. 2001; 280:196–203. [DOI] [PubMed] [Google Scholar]
- 38. Zhao C, Nie H, Shen Q, Zhang S, Lukowitz W, Tang D. EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 2014; 10:e1004389 10.1371/journal.pgen.1004389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao L, Xiang CB. The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol Biol. 2008; 67:125–134. 10.1007/s11103-008-9306-8 [DOI] [PubMed] [Google Scholar]
- 40. Huang Y, Li CY, Qi Y, Park S, Gibson SI. SIS8, a putative mitogen-activated protein kinase kinase kinase, regulates sugar-resistant seedling development in Arabidopsis. Plant J. 2014; 77: 577–588. 10.1111/tpj.12404 [DOI] [PubMed] [Google Scholar]
- 41. Sasayama D, Matsuoka D, Oka M, Shitamichi N, Furuya T, Azuma T, et al. MAP3Kδ4, an Arabidopsis Raf-like MAP3K, regulates plant growth and shoot branching. Plant Biotechnol. 2011; 28:463–470. [Google Scholar]
- 42. Ning J, Li X, Hicks LM, Xiong L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 2010; 152:876–890. 10.1104/pp.109.149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ning J, Zhang B, Wang N, Zhou Y, Xiong L. Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell. 2011; 23: 4334–4347. 10.1105/tpc.111.093419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shen X, Liu H, Yuan B, Li X, Xu C, Wang S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell. 2011; Environ 34:179–191. [DOI] [PubMed] [Google Scholar]
- 45. Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 46. Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006; 18:257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schonfeld MA, Johnson RC, Carver BF. Water relations in winter wheat as drought resistance indicator. Crop Sci. 1998; 28: 526–531. [Google Scholar]
- 48. Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, et al. T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 2002; 3:1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mehrotra R, Bhalothia P, Bansal P, Basantani MK, Bharti V, Mehrotra S. Abscisic acid and abiotic stress tolerance—different tiers of regulation. J Plant Physiol. 2014; 171:486–496. 10.1016/j.jplph.2013.12.007 [DOI] [PubMed] [Google Scholar]
- 50. Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000; 123: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001; 26: 421–433. [DOI] [PubMed] [Google Scholar]
- 52. Gonzalez-Guzman M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002; 14:1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gilmour SJ, Artus NN, Thomashow MF. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana . Plant Mol Biol. 1992; 18: 13–21. [DOI] [PubMed] [Google Scholar]
- 54. Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaquchi-Shinozaki K, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998; 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shitamichi N, Matsuoka D, Sasayama D, Furuya T, Nanmori T. Over-expression of MAP3Kδ4, an ABA-inducible Raf-like MAP3K that confers salt tolerance in Arabidopsis. Plant Biotechnol. 2013; 30:111–118. [Google Scholar]
- 56. Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, et al. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006; 9:436–442. [DOI] [PubMed] [Google Scholar]
- 57. Menges M, Dóczi R, Okrész L, Morandini P, Mizzi L, Soloview M, et al. Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol. 2008; 179:643–662. 10.1111/j.1469-8137.2008.02552.x [DOI] [PubMed] [Google Scholar]
- 58. Shou H, Bordallo P, Fan JB, Yeakley JM, Bibikova M, Sheen J, et al. Expression of an active tobacco mitogen-activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proc Natl Acad Sci USA. 2004; 101: 3298–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shou H, Bordallo P, Wang K. Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J Exp Bot. 2004; 55:1013–1019. [DOI] [PubMed] [Google Scholar]
- 60. Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ. Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 2003; 33: 221–233. [DOI] [PubMed] [Google Scholar]
- 61. Tang D, Innes RW. Overexpression of a kinase-deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene-induced senescence in Arabidopsis. Plant J. 2002; 32:975–983. [DOI] [PubMed] [Google Scholar]
- 62. Furuya T, Matsuoka D, Nanmori T. Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. J Plant Res. 2013; 126:833–840. 10.1007/s10265-013-0576-0 [DOI] [PubMed] [Google Scholar]
- 63. Doczi R, Brader G, Pettkó-Szandtner A, Rajh I, Djamei A, Pitzschke A, et al. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell. 2007; 19: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li X, Zhang Y, Huang L, Ouyang Z, Hong Y, Zhang H, et al. Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea . BMC Plant Biol. 2014; 14:166 10.1186/1471-2229-14-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002; 415: 977–983. [DOI] [PubMed] [Google Scholar]
- 66. Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007; 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 2003; 17: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Miao Y, Laun TM, Smykowski A, Zentgraf U. Arabidopsis MEKK1 can take a short cut: It can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol. 2007; 65: 63–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.