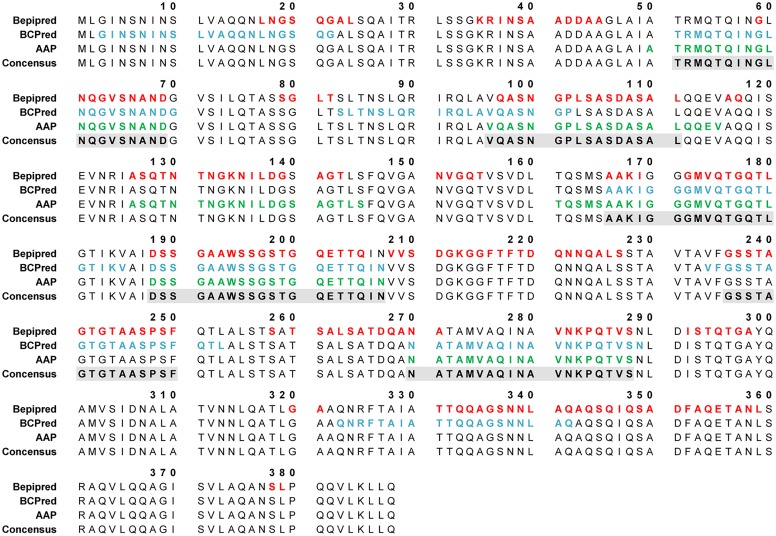
Fig 1. FliCBp B-cell linear sequence-based epitope predictions using three predictor methods: BepiPred (threshold = 0.35), BCPred (classifier specificity = 75%), and AAP (classifier specificity = 75%).
Epitope predictions were carried out on the full-length amino acid sequence of FliCBp, using three web-accessible prediction servers: BepiPred, BCPred and AAP [21,22]. Predicted epitope residues by BepiPred, BCPred and AAP are shown in red, blue and green font, respectively on the amino acid sequence (residue numbers are indicated). Grey shaded boxes indicate consensus positive residues identified by at least two epitope predictors.