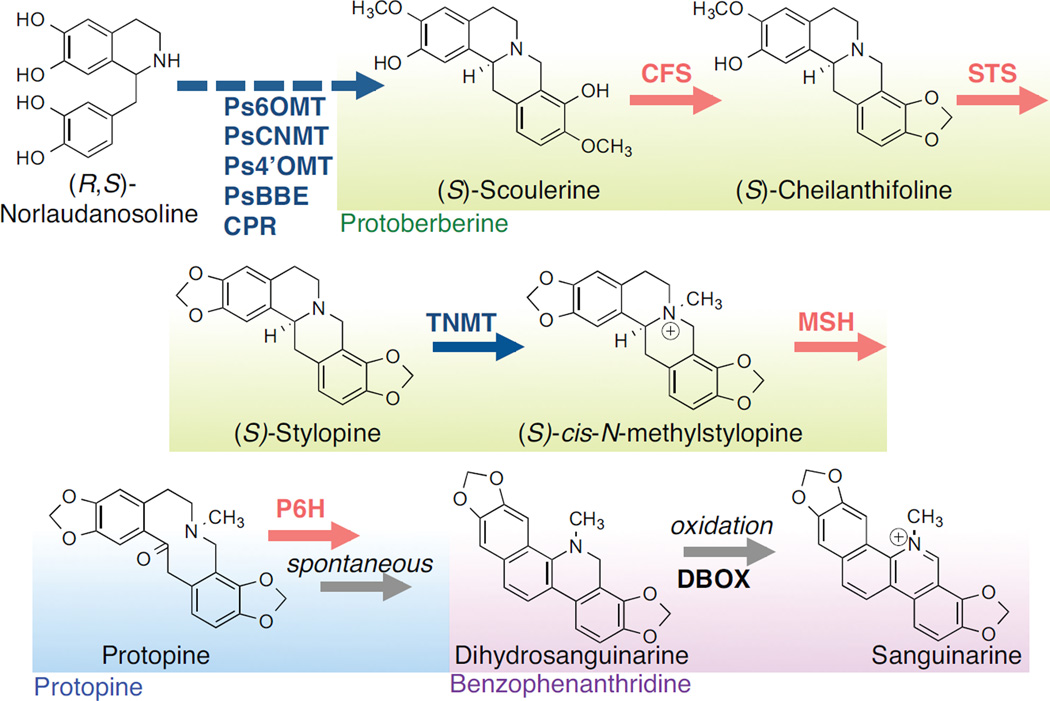
Figure 1.
Engineered pathway for sanguinarine biosynthesis in yeast from (R, S)-norlaudanosoline. Red arrows indicate reactions catalyzed by a plant cytochrome P450 enzyme. Structural class of the metabolites are indicated as green, protoberberine; blue, protopine; purple, benzophenanthridine. Ps6OMT, P. somniferum 6-O-methyltransferase; PsCNMT, P. somniferum coclaurine N-methyltransferase; Ps4’OMT, P. somniferum 4’-O-methyltransferase; PsBBE, P. somniferum berberine bridge enzyme; CPR, cytochrome P450 reductase; CFS, cheilanthifoline synthase; STS, stylopine synthase; TNMT, tetrahydroprotoberberine N-methyltransferase; MSH, cis-N-methylstylopine 14-hydroxylase; P6H, protopine 6-hydroxylase; DBOX, dihydrobenzophenanthridine oxidase.