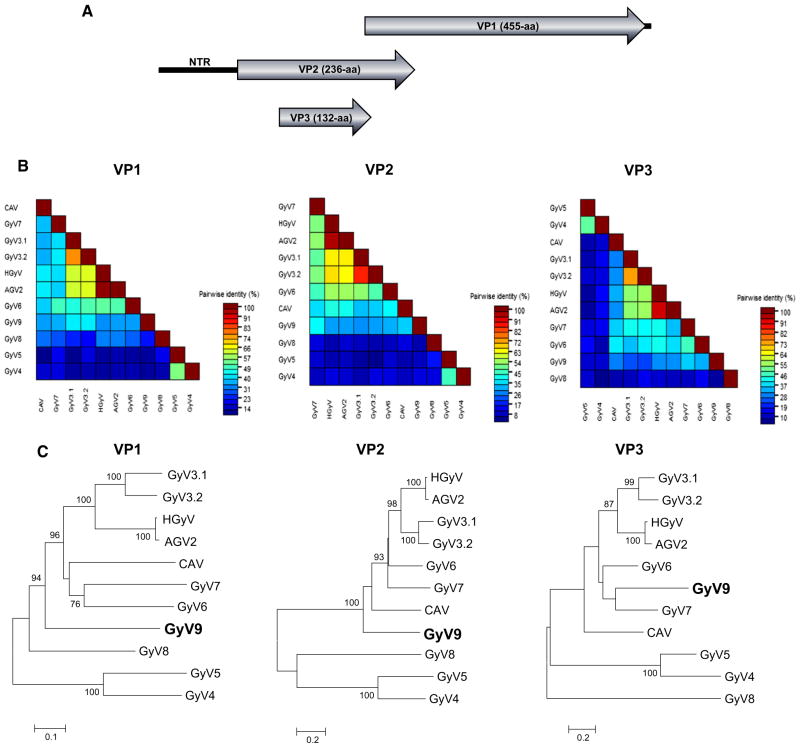
Fig. 1.
New gyrovirus genome and phylogeny. a Organization of the circular DNA gyrovirus 9 genome shown here in a linear form. b Pairwise comparison of VP1, VP2, and VP3 proteins of the new gyrovirus and other species in the Gyrovirus genus. c Phylogenetic trees generated with VP1, VP2, and VP3 proteins of the new gyrovirus and other species in the Gyrovirus genus. The scale indicated amino acid substitutions per position. Bootstrap values (based on 100 replicates) for each node are given if >70. CLUSTAL X with the default settings included gap opening penalty (10), gap extension penalty (0.2), protein weight matrix (gonnet), residue-specific penalities (on), hydrophilic penalties (on), gap separation distance (4), and end gap separation (off)