Abstract
Endoplasmic reticulum stress (ERS), which is a novel pathway of regulating cellular apoptosis and the function of ERS during corpus luteum (CL) regression, is explored. Early-luteal stage (day 2), mid-luteal stage (day 7), and late-luteal stage (day 14 and 20) were induced, and the apoptosis of luteal cells was detected by a terminal 2′-deoxyuridine 5′-triphosphate nick-end labeling (TUNEL) assay. The apoptotic cells were increased with the regression of CL, especially during the late-luteal stage. The ERS markers glucose-regulated protein 78 (Grp78), CCAAT/enhancer-binding protein homologous protein (CHOP), X-box binding protein 1 (XBP1), activating transcription factor 6α (ATF6α), eukaryotic initiation factor 2α (eIF2α), inositol-requiring protein 1α (IRE1α), caspase 12, and apoptosis marker caspase 3 were analyzed by real-time polymerase chain reaction (PCR) and immunohistochemistry, in agreement with the results of the TUNEL assay; the expression levels of CHOP, caspase 12, and caspase 3 were increased during the process of CL regression. Luteal cells were isolated and cultured in vitro, and the apoptosis of luteal cells was induced by prostaglandin F2α. The ERS was attenuated by the ERS inhibitor tauroursodeoxycholic acid, and the apoptotic rate was analyzed by flow cytometry. The ERS markers Grp78, CHOP, XBP1s, ATF6α, eIF2α, IRE1α, caspase 12, and apoptotic execute marker caspase 3 were analyzed by real-time PCR and immunofluorescence, and the results suggested that the expression of CHOP, caspase 12, and caspase 3 were increased, and there was increased apoptosis of luteal cells. But the expression of IRE1α/XBP1s and eIF2α was not detected. Taken together, the ERS is involved in the CL regression of rats through the CHOP and caspase 12 pathway.
Keywords: endoplasmic reticulum stress, corpus luteum regression, rats, apoptosis
Introduction
The corpus luteum (CL) is a temporary, rhythmic endocrine gland that develops after ovulation from the ruptured follicle during the luteal phase. Progesterone is produced by the CL. Progesterone is indispensable for the establishment and maintenance of intrauterine pregnancies in mammals.1–3 Several studies revealed that the function and fate of the CL are mediated by a number of factors, including steroids, growth factors, gonadotropins, prostaglandins, cytokines, and peptide hormones,4 but the accurate controlling mechanism of regression of the CL is unknown. Previous studies suggested that cellular apoptosis was involved in the regression of CL,5 and apoptosis was detected in many species during spontaneous and induced CL regression, including cows,6,7 rats,8–10 sheep,11 and humans.12 Previous studies suggested that the death receptor- and mitochondria-mediated apoptotic systems have been shown to be active in regression of the CL.13–15
Endoplasmic reticulum stress (ERS) is a novel pathway of cellular apoptosis and generally occurs in secretory cell types.16 78-kDa Glucose-regulated protein (Grp78) is an endoplasmic reticulum chaperone, with multiple functional roles in protein processing and provision of cellular protection.17 Although the ERS is initiated and the increased Grp78 is relocalized from the endoplasmic reticulum (ER), the signaling regulator Grp78 is used as a monitor of ERS.18
In the early stages, ERS is alleviated by the unfold protein response (UPR). The following 3 signal transduction pathways are involved in the UPR: protein kinase RNA-like ER kinase (PERK), inositol-requiring enzyme-1/X-box-binding protein (IRE1/XBP-1), and activating transcription factor 6 (ATF6). If the ERS is not mitigated through the UPR, persistent and severe ERS results in cellular apoptosis. The apoptosis was initiated by upregulating the ERS-associated proapoptotic marker CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP/GADD153), by the ER-associated caspase 12 pathway and by activation of the c-Jun NH2-terminal kinase (JNK)-dependent pathway.19–21 To date, the accurate signal pathway of ERS-mediated apoptosis was still fully understood. Previous study suggested that the apoptotic response is mediated through activation of CHOP primarily by signaling of the activating transcription factor 6α (ATF6α) and the PERK/eukaryotic initiation factor 2α (eIF2α)/ATF4.22 Indeed, CHOP was target gene of 3 UPR signal pathways23 and proapoptosis during ERS response.24
Tauroursodeoxycholic acid (TUDCA) is an endogenous hydrophilic bile acid with multiple properties,25 and recent studies have shown that TUDCA functions as chemical chaperone and helps attenuate ERS.26
Previous studies suggested that ERS was involved in several important reproductive process, including development of preimplantation embryo,27–31 follicle atresia,32,33 embryonic implantation,34 decidualization,35 and placentation.36–41 Whether ERS is involved in the regression of the CL is unknown. Therefore, in this study, the function of ERS in regression of the CL is analyzed.
Materials and Methods
Animal Treatment
Immature female rats were obtained from the Experimental Animal Center of Ningxia Medical University. The rats were housed in a controlled temperature and humidity environment with a 12-hour light–dark cycle, and they were fed a standard diet. The rats had access to food and water ad libitum, and all procedures were approved by the Animal Care and Use committee at Ningxia Medical University.
Immature (26-day-old) female rats were superovulated by an intraperitoneal (ip) injection of 50 IU Pregnant Mare Serum Gonadotrophin (PMSG), (Sansheng pharmaceutical Co, Ltd, Ningbo, China). After 48 hours, the rats were injected with 50 IU of Human Chorionic Gonadotropin (HCG) (Sansheng pharmaceutical Co, Ltd) ip to induce ovulation and pseudopregnancy. The rats were killed by cervical dislocation, and the ovaries were excised on days 2, 7, 14, and 20 after hCG administration, 10 rats in each group. These time points represent the early-luteal stage (day 2), mid-luteal stage (day 7), and late-luteal stage (day 14 and 20).3,42 The CLs dissected under microscope from left ovaries were used for RNA extraction, and the right ovaries were used for immunohistochemistry.
Real-Time Polymerase Chain Reaction
Total RNA was extracted using the Trizol reagent (TaKaRa, Dalian, China) according to the manufacturer’s instructions, and the purity of the RNA was verified according to previous description.33 The complementary DNA was reverse transcribed using the PrimeScipt RT reagent kit (TaKaRa) according to the manufacturer’s instructions.
Real-time polymerase chain reaction (PCR) was carried out using an ABI 7500 fast machine and 7500 Software v2.0.5 (7500 Fast Real-Time PCR System, ABI, Grand Island, NY). The mouse GAPDH gene, which was used as the reference gene, was amplified in parallel with the target gene to allow for gene expression normalization, and the 2−△△ Ct was used for quantification. The detection of real-time PCR products was performed using the SYBR Premix Ex Taq II (TaKaRa) following the manufacturer’s instructions. Each PCR reaction was performed in a 20.0 μL reaction mixture containing 10.0 μL of 2 × SYBR Premix Ex Taq II, 2.0 of μL cDNA (the equivalent of 20 ng total RNA) as the template, 0.8 μL of each primer at 10 μmol/L, and 6.4 μL of nuclease-free water. The PCR cycling conditions comprised 1 cycle at 95°C for 30 seconds, followed by 40 cycles at 95°C for 5 seconds, and at 60°C for 30 seconds. The experiments were performed in triplicate for each data point, and the mean of all the values was used for the final analysis. The primers are shown in Table 1.
Table 1.
Primer Sequences Used for Real-Time Quantitative PCR.
| Target gene | GenBank Accession No. | Primer Sequence | Product Size, bp | Annealing Temperature, °C |
|---|---|---|---|---|
| XBP1s | NM_001271731.1 | F:5′-GGTCTGCTGAGTCCGCAGCAGG-3′ R:5′-GAAAGGGAGGCTGGTAAGGAAC-3′ |
232 | 60 |
| ATF6α | NM_001107196.1 | F: 5′-GGG AGT GAG CTG CAG GTG TA-3′ R: 5′-TTA TGG GTG GTA GCT GGT AA-3′ |
137 | 60 |
| eIF2α | NM_019356.1 | F:5′-GCTTGCTATGGTTACGAAGGC-3′ R:5′-CATCACATACCTGGGTGGAG-3′ |
120 | 60 |
| IRE1α | NM_001191926.1 | F: 5′-CGA GCC ATG AGGAAT AAG AG-3′ R: 5′-GGA AAC GTG ATG TGA AGT AG-3′ |
109 | 60 |
| Grp78 | NM_013083.2 | F:5′-AGAAACTCCGGCGTGAGGTAGA-3′ R:5′-TTTCTGGACAGGTTTCATGGTAG-3′ |
176 | 60 |
| CHOP | NM_024134.2 | F: 5′-AGCTGGAAGCCTGGTATGAGGA-3′ R: 5′-AGCTAGGGATGCAGGGTCAA-3′ |
134 | 60 |
| Caspase 12 | NM_130422.1 | F:5′-CTGGCCCTCATCATCTGCAACAA-3′ R: 5′-CGGCCAGCAAACTTCATTAAT-3′ |
173 | 60 |
| Caspase 3 | NM_012922.2 | F: 5′-ACTGGAAAGCCGAAACTCTTC-3′ R: 5′-AGTTCCACTGTCTGTCTCAATA-3′ |
81 | 60 |
| GAPDH | NM_017008.4 | F: 5′-GGCATCGTGGAAGGGCTC-3′ R: 5′-GACCTTGCCCACAGCCTT-3′ |
156 | 60 |
Abbreviations: PCR, polymerase chain reaction; XBP1, X-box binding protein 1; ATF6α, activating transcription factor 6α; eIF2α, eukaryotic initiation factor 2α; IRE1α, inositol-requiring protein 1α; Grp78, glucose-regulated protein 78; CHOP, CCAAT/enhancer-binding protein homologous protein.
Immunohistochemistry
The mouse ovaries were collected and immediately fixed with 4% paraformaldehyde for 24 hours. The preparation of the paraffin-embedded ovaries was performed according to the standard procedure, and the embedded ovaries were cut into 6-μm sections. The sections were dehydrated at 37°C overnight. The antigen heating recovery with a mixed liquid of 0.1mol/L citric acid, 0.1 molL natrium citricum, and distilled water (9:41:450) was performed for 20 minutes at 96°C to 98°C, and the solution was cooled for 20 minutes at room temperature. The immunohistochemistry (IHC) experiments were performed using the Histostain-Plus Kit (Beijing 4A Biotech Co, Ltd, China), and the process was performed according to the manufacturer’s instructions. The sections were treated with 0.3% H2O2–methanol for 10 minutes to block the endogenous peroxidase activity and washed in 0.01 mol/L phosphate-buffered saline (PBS) 3 times. Nonspecific binding was blocked in 10% normal goat serum in PBS for 1 hour. The sections were incubated with an anti-Grp78 rabbit polyclonal antibody (sc-13968, 200 μg/mL, Santa Cruz Biotechnology Inc, Dallas, Texas), an anti-CHOP mouse polyclonal antibody (sc-7351, 200 μg/mL, Santa Cruz Biotechnology Inc), an anti-ATF6α rabbit polyclonal antibody (sc-22799, 200 μg/mL, Santa Cruz Biotechnology Inc), an anticaspase 12 rabbit polyclonal antibody (sc-5627, 200 μg/mL, Santa Cruz Biotechnology Inc, Littleton, Colorado), anti-p(ser724) IREα (nb100-2323, 1:100 dilution; Novus Biologicals, Colorado), and an antiactive caspase 3 rabbit polyclonal antibody (ab2302, 1:100 dilution, United Kingdom) at 37°C (60 minutes). The sections were then washed in 0.01 mol/L PBS 3 times and incubated (30 minutes) with a biotinylated goat antirabbit or goat antimouse secondary antibody at 37°C. The slides were rinsed, incubated in streptavidin-horseradish peroxidase for 10 minutes at 37°C, rinsed again, and incubated (5-10 minutes) in 3,3′-diaminobenzidine tetrahydrochloride chromogen as the substrate. After a final rinse with ddH2O, the sections were counterstained with hematoxylin, ethanol dehydrated, and mounted using neutral balsam.
Apoptosis/Terminal Transferase 2′-deoxyuridine 5′-triphosphate Nick End Labeling Assay
The paraffin-embedded sections were treated with dimethylbenzene for 5 minutes twice, hydrated with gradient alcohol for 5 minutes (100%, 100%, 95%, 85%, 70%, and 50%), washed in PBS for 5 minutes 3 times, treated with 4% (v/v) paraformaldehyde for 15 minutes, washed with PBS for 5 minutes twice, treated with proteinase K for 10 minutes, washed in PBS for 5 minutes twice, treated with 4% (v/v) paraformaldehyde for 5 minutes, and washed in PBS for 5 minutes twice. Apoptotic cells were detected by the TUNEL (TdT mediated dUTP nick-end labeling) method according to the kit’s instruction (Promega Corp, Madison, Wisconsin). Briefly, 100 μL of equilibrium buffer was added to the sections at room temperature for 10 minutes. The equilibrium buffer was discarded, and 50 μL of rTdT was added. The sections were incubated at 37°C for 60 minutes. After incubation, the reaction was terminated with 2 × SSC at room temperature for 15 minutes. The sections were washed with PBS for 5 minutes 3 times, and the nuclei were counterstained with propidium iodide (PI) for 5 minutes and washed with PBS for 5 minutes 3 times. The fluorescence of the sections was photographed by fluorescence microscopy.
Isolation and culture of luteal cell
Luteal cells were isolated from rat ovaries on day 7 of pseudopregnancy according to a previous description with minor modifications.3 Briefly, the CLs were digested with 0.25% trypsin-EDTA at 37°C for 10 minutes. The reaction was terminated with Dulbecco Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS) and filtered through a 200-mm Falcon nylon mesh (NO.RA442, Shanghai Sangon Biotech Co, Ltd, Shanghai, China) to remove debris. The filtrate was moved into a 1.5-mL centrifuge tube and centrifuged at 1000 rpm for 5 minutes. The supernatant was discarded, and the precipitate was washed with 0.01 mol/L PBS and then washed with DMEM/F12 supplemented with 10% FBS twice. The collected cells were seeded at a concentration of 1 × 106 cells/mL in culture dishes and incubated at 37°C with 5% CO2 in DMEM/F12 supplemented with 10% FBS, 100 U/mL penicillin/mL, and 100 mg/mL streptomycin (Gibco BRL, Grand Island). After 72 hours of incubation, the medium and unattached cells were removed, and the cells divided into the following 4 groups: (1) addition of basic serum-free medium (DMEM/F12) for serum starvation; (2) addition of DMEM/F12 medium with 100 μmol/L prostaglandin F2α (PGF2α; 100 μmol/L; Ningbo Sansheng Co., Ltd. Ningbo, China) to induce CL regression; (3) addition of DMEM/F12 medium with 10 mmol/L TUDCA for attenuating the ERS and cell death; (4) addition of DMEM/F12 medium with 100 μmol/L PGF2α plus 10 mmol/L TUDCA. After 24 hours, the cells were collected for real-time PCR, confocal laser scanning microscopy (CLSM), and flow cytometry detection.
Confocal Laser Scanning Microscopy
The apoptotic luteal cells were detected through colocalization of the ERS-related apoptotic marker CHOP and caspase 12 by a CLSM. The process of CLSM was performed according to a previous description.33 Briefly, the luteal cells were cultured in the CLSM culture dishes and divided into 4 groups. After 24 hours of treatment, the luteal cells were immediately fixed in 4% paraformaldehyde for 20 minutes. After fixation, the apoptotic luteal cells were washed in 0.01 mol/L PBS for 3 minutes and blocked in 10% normal goat serum for 30 minutes. After blocking, the cells were incubated overnight with a mouse anti-CHOP polyclonal antibody, a rabbit anticaspase 12 polyclonal antibody at 4°C, and washed with PBS for 5 minutes 3 times. The cells were incubated with fluorescein-conjugated AffiniPure Goat Anti-Rabbit IgG (Green, ZF-0311, ZSGB-BIO, Beijing, China) and rhodamine (tetramethylrhodamine isothiocyanate)-conjugated AffiniPure Goat Anti-mouse IgG (Red, ZF-0313, ZSGB-BIO, Beijing, China). The cells were washed with PBS for 5 minutes 3 times, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole for 5 minutes at room temperature. The cells were washed with PBS for 5 minutes 3 times, and the culture dishes were transferred into CLSM and photographed.
Detection of Apoptotic Rate of Luteal Cells
To detect the apoptotic rate of the luteal cells in the 4 experimental groups, the Annexin V-fluorescein isothiocyanate (FITC) Assay Kit (BB-4101-50t, Bestbio, Shanghai, China) was used according to the manufacturer’s instructions. Briefly, the luteal cells were washed twice with cold PBS, and the PBS was gently removed from the cell pellet. The cells were resuspended in 500 μL of binding buffer with 5 μL annexin V-FITC and 5 μL of propidium iodide added. The cells were gently vortexed and incubated for 15 minutes in the dark. Detection by flow cytometry (EPICS Altra, Beckman Coulter Cytomics Altra, Brea, California) was performed within 1 hour. The apoptotic cells were determined by counting the percentage of AnnexinV-FITC+/PI- and AnnexinV-FITC+/PI+ cells.
Statistical Analysis
All experiments were replicated at least 3 times for each group, and the data were presented as the means ± standard error of the mean. The data were analyzed with 1-way analysis of variance followed by post hoc test performed by Fisher least significant difference test with SPSS software (Version 13.0; SPSS, Inc., Chicago, Illinois). Differences were considered significant when P < .05.
Results
The Detection of Apoptotic Luteal Cells During CL Regression by TUNEL
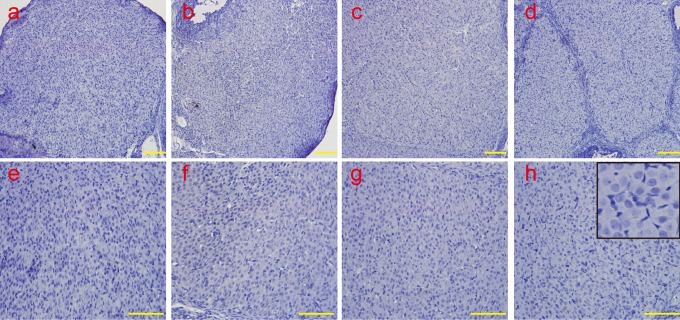
The apoptotic luteal cells during the early-luteal stage, mid-luteal stage, and late-luteal stage were analyzed by TUNEL, and the results suggested that there were very few apoptotic luteal cells in the early-luteal stage (Figure 1A). The apoptotic luteal cells gradually increased in the midluteal stage (Figure 1B) and were remarkably increased in the late-luteal stage (Figure 1C and D).
Figure 1.
The detection of cellular apoptosis by TUNEL during the CL regression. Very few apoptotic luteal cells were found in the early-luteal stage, and the number gradually increased in the mid-luteal stage; the number was remarkably increased in the late-luteal stage. a, The early-luteal stage (2 d); b, the mid-luteal stage (7 d); c, the late-luteal stage (14 d); and d, the late-luteal stage (20 d). Scale bar: a, b, c, and d: 200 μm. TUNEL indicates terminal 2′-deoxyuridine 5′-triphosphate nick-end labeling; CL, corpus luteum; d, day.
The Detection of ERS-Related and Apoptotic Execution Molecules by Real-Time PCR
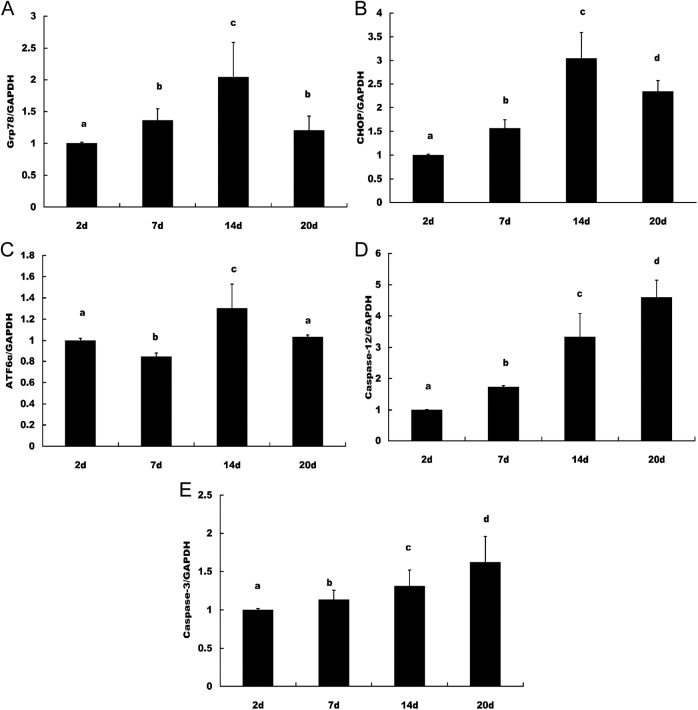
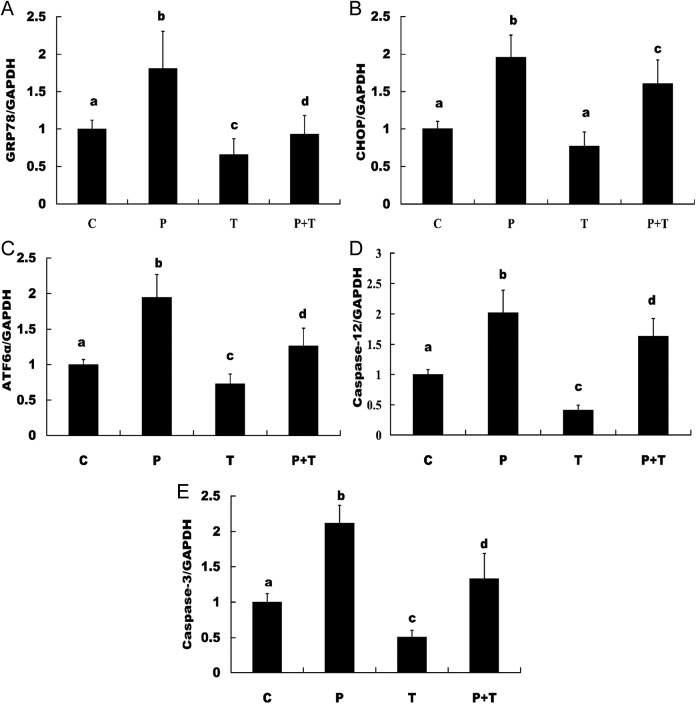
The ERS markers Grp78, CHOP, XBP1s, ATF6α, eIF2α, IRE1α, caspase 12, and apoptotic execution marker caspase 3 were checked by real-time PCR. The results suggested that Grp78 and CHOP increased from the early luteal stage and peaked in the late-luteal stage on day 14 and decreased on day 20; ATF6α decreased in the early luteal stage on day 7 compared with day 2, but peaked in the late-luteal stage on day 14 and decreased on day 20. The apoptotic marker caspase 12 and apoptotic execution marker caspase 3 increased from the early-luteal stage and peaked in the late-luteal stage on day 20 (Figure 2). The results from the in vitro cultured and treated luteal cells suggested that the expression of Grp78, CHOP, ATF6α, caspase 12, and caspase 3 was detectable and significantly increased in the luteal cells treated with PGF2α and significantly decreased in the luteal cells treated with TUDCA compared with the control group and the group treated with TUDCA plus PGF2α (Figure 3). The expression of eIF2α, IRE1α, and XBP1s was not detected.
Figure 2.
ERS and apoptotic molecules in induced CL regression by real-time PCR The expression levels of Grp78, CHOP, and ATF6α were increased from the early-luteal stage and peaked in the late-luteal stage on day 14, but the levels were decreased on day 20. The apoptotic marker caspase 12 and apoptotic execution marker caspase 3 were increased from the early-luteal stage and peaked in the late-luteal stage on day 20. A, Grp78, (B) CHOP, (C) ATF6α, (D) caspase 12, and (E) caspase 3. The different letters (a, b, c, and d) indicate significant differences, and the same letters indicate no difference. ERS indicates endoplasmic reticulum stress; CL, corpus luteum; PCR, polymerase chain reaction; Grp78, glucose-regulated protein 78; CHOP, CCAAT/enhancer-binding protein homologous protein; ATF6α, activating transcription factor 6α.
Figure 3.
Examination of ERS and apoptotic molecules in cultured luteal cells in vitro by real-time PCR. The expression of Grp78, CHOP, ATF6α, caspase 12, and caspase-3 was examined and found to be significantly increased in the luteal cells treated with PGF2α, and expression of these molecules was significantly decreased in the luteal cells treated with TUDCA compared to the control group and the group treated with TUDCA plus PGF2α. A, Grp78, (B) CHOP, (C) ATF6α, (D) caspase 12, and (E) caspase 3. The different letters (a, b, c, and d) indicate significant differences, and the same letters indicate no difference. ERS indicates endoplasmic reticulum stress; PCR, polymerase chain reaction; Grp78, glucose-regulated protein 78; CHOP, CCAAT/enhancer-binding protein homologous protein; ATF6α, activating transcription factor 6α; PGF2α, prostaglandin F2α; TUDCA, tauroursodeoxycholic acid.
Analysis of the ERS-Related and Apoptotic Execution Molecules by IHC
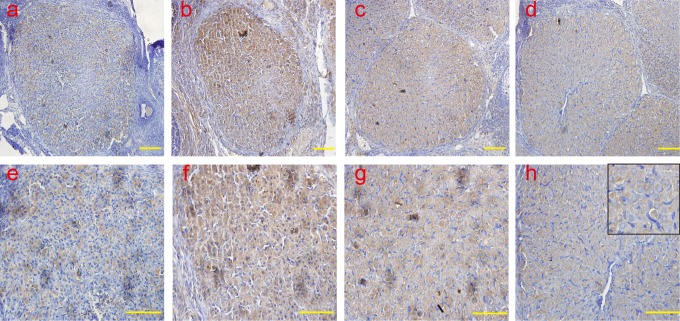
The protein expression and localization of Grp78, CHOP, ATF6α, IRE-1, caspase 12, and active caspase 3 were examined by IHC, and it was observed that Grp78 was mainly localized in the cytoplasm, and CHOP, ATF6α, and caspase12 proteins were detected in the cytoplasm and nucleus. Active caspase 3 was mainly localized in the nucleus. Consistent with the results of the real-time PCR, the protein expression of Grp78 (Figure 4), CHOP (Figure 5), ATF6α (Figure 6), and caspase-12 (Figure 7) was detected, and the peak of Grp78 was found in the late-luteal stage on day 14; the level decreased on day 20. Levels of CHOP, ATF6α, and caspase-12 were remarkably increased in the late-luteal stage on days 14 and 20. The active caspase 3 (Figure 8) protein was barely detectable in the early-luteal stage and mid-luteal stage and remarkably increased in the late-luteal stage; the level peaked on day 20. In accordance with the results of the real-time PCR, the IRE-1 (Figure 9) protein was not found during the early-luteal stage, mid-luteal stage, or late-luteal stage.
Figure 4.
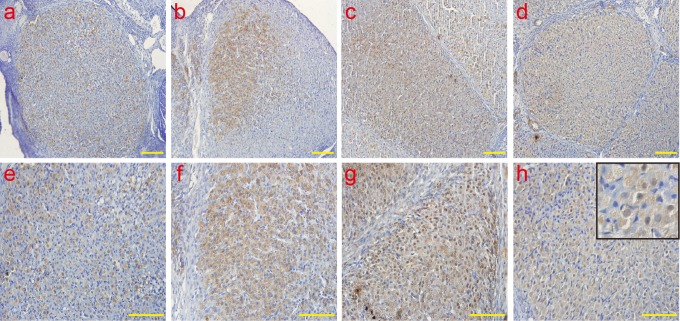
The analysis of ERS molecule Grp78 by IHC Grp78 was mainly localized in the cytoplasm. Consistent with the results of the real-time PCR, the protein expression of Grp78 was examined, and the peaks of Grp78 emerged in the late-luteal stage on day 14; the levels decreased on day 20. a and e: early-luteal stage (2 d); b and f: mid-luteal stage (7 d); c and g: late-luteal stage (14 d); d and h: late-luteal stage (20 d). The picture in the upper right corner of h indicates the enlargement. Scale bar: a, b, c, and d: 50 μm; e, f, g, and h: 100 μm. ERS indicates endoplasmic reticulum; Grp78, glucose-regulated protein 78; IHC, immunohistochemistry; Grp78, glucose-regulated protein 78; PCR, polymerase chain reaction; d, day.
Figure 5.
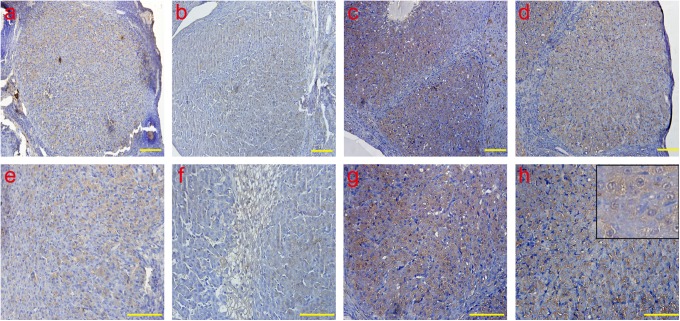
The analysis of ERS-associated proapoptotic molecular CHOP by IHC. The CHOP protein was detected in the cytoplasm and the nucleus. Consistent with the results of the real-time PCR, the protein expression of CHOP was examined, and the peaks of CHOP emerged in the late-luteal stage on day 14; the levels decreased on day 20. a and e: early-luteal stage (2 d); b and f: mid-luteal stage (7 d); c and g: late-luteal stage (14 d); d and h: late-luteal stage (20 d). The picture in the upper right corner of h indicates the enlargement. Scale bar: a, b, c and, d: 50 μm; e, f, g, and h: 100 μm. ERS indicates endoplasmic reticulum; CHOP, CCAAT/enhancer-binding protein homologous protein; IHC, immunohistochemistry; PCR, polymerase chain reaction; d, day.
Figure 6.
The analysis of ERS molecules ATF6α by IHC. The ATF6α protein was detected in the cytoplasm and the nucleus. Consistent with the results of the real-time PCR, the protein expression of ATF6α was examined, and the peaks of ATF6α emerged in the late-luteal stage on day 14; the levels decreased on day 20. a and e: early-luteal stage (2 d); b and f: mid-luteal stage (7 d); c and g: late-luteal stage (14 d); d and h: late-luteal stage (20 d). The picture in the upper right corner of h indicates the enlargement. Scale bar: a, b, c, and d: 50 μm; e, f, g, and h: 100 μm. ERS indicates endoplasmic reticulum; ATF6α, activating transcription factor 6α; IHC, immunohistochemistry; PCR, polymerase chain reaction; d, day.
Figure 7.
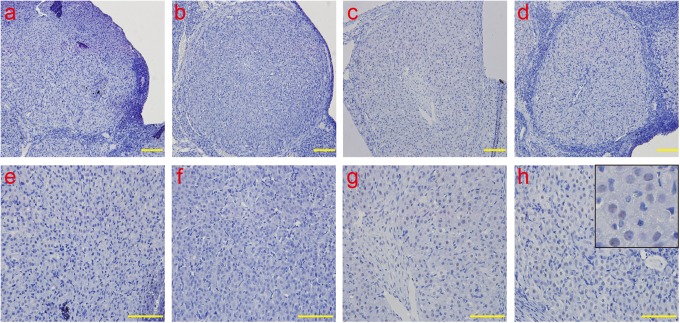
The analysis of ERS-associated apoptotic molecular caspase 12 by IHC. The caspase-12 protein was detected in the cytoplasm and the nucleus. Consistent with the results of the real-time PCR, the caspase-12 protein was weakly detected in the early-luteal stage and mid-luteal stage and remarkably increased in the late-luteal stage; the levels peaked on day 20. a and e: early-luteal stage (2 d); b and f: mid-luteal stage (7 d); c and g: late-luteal stage (14 d); d and h: late-luteal stage (20 d). The picture in the upper right corner of h indicates the enlargement. Scale bar: a, b, c, and d: 50 μm; e, f, g, and h: 100 μm. ERS indicates endoplasmic reticulum; IHC, immunohistochemistry; PCR, polymerase chain reaction; d, day.
Figure 8.
The analysis of apoptotic execute molecular active caspase 3 by IHC. Active caspase 3 was mainly localized in the nucleus. Consistent with the results of the real-time PCR, the active caspase-3 protein was weakly detected in the early-luteal stage and mid-luteal stage and remarkably increased in the late-luteal stage; the levels peaked on day 20. a and e: early-luteal stage (2 d); b and f: mid-luteal stage (7 d); c and g: late-luteal stage (14 d); d and h: late-luteal stage (20 d). The picture in the upper right corner of h indicates the enlargement. Scale bar: a, b, c, and d: 50 μm; e, f, g, and h: 100 μm. IHC indicates immunohistochemistry; PCR, polymerase chain reaction; d, day.
Figure 9.
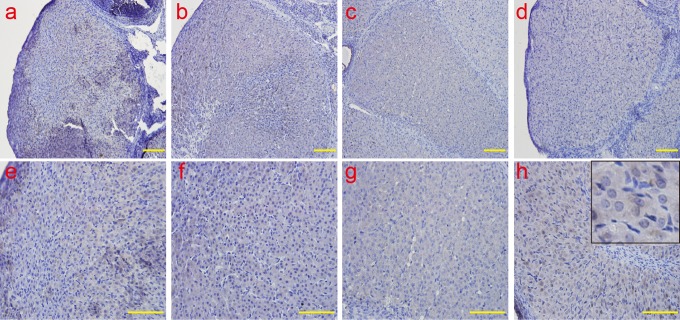
The analysis of ERS molecules IRE1α by IHC. Consistent with the results of the real-time PCR, the protein expression of IRE-1 was not detected during all stages by IHC. a and e: early-luteal stage (2 d); b and f: mid-luteal stage (7 d); c and g: late-luteal stage (14 d); d and h: late-luteal stage (20 d). The picture in the upper right corner of h indicates the enlargement. Scale bar: a, b, c, and d: 50 μm; e, f, g, and h: 100 μm. ERS indicates endoplasmic reticulum; IRE1α, inositol-requiring protein 1α; IHC, immunohistochemistry; PCR, polymerase chain reaction; d, day.
The Analysis of Luteal Cell Apoptosis
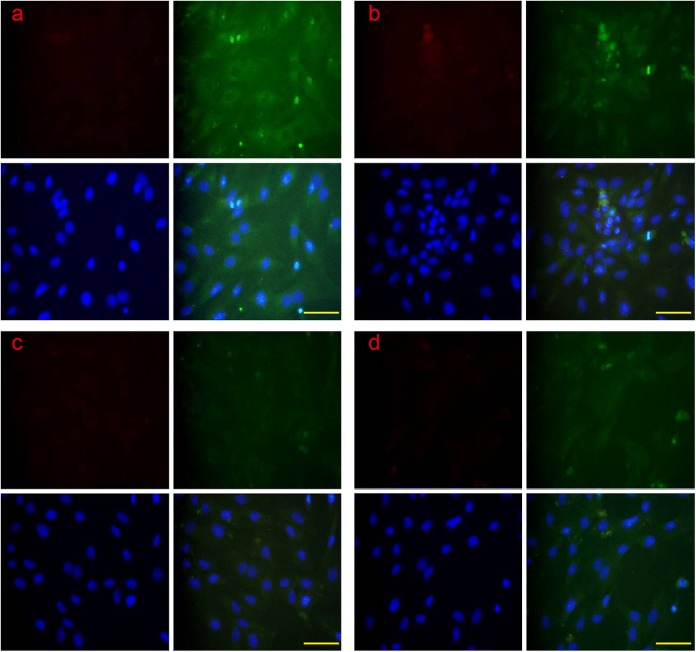
Apoptosis in the 4 groups of luteal cell was examined by CLSM through colocalization of the RES-related proapoptotic protein CHOP and the apoptotic protein caspase 12; the evaluative criteria of early apoptosis and late apoptosis were according a previous description.33 The results suggested that the CHOP and caspase-12 proteins were all found in early apoptosis and late apoptosis. The CHOP and caspase-12 proteins were translocated from the cytoplasm into the nucleus during late apoptosis (Figure 10). The apoptotic rate of the luteal cells was examined by flow cytometry, and the results suggested that the apoptotic rate of the group treated with PGF2α was significantly higher than the other groups, but the apoptotic rate of the group treated with TUDCA was significantly lower than the other groups (Figure 11).
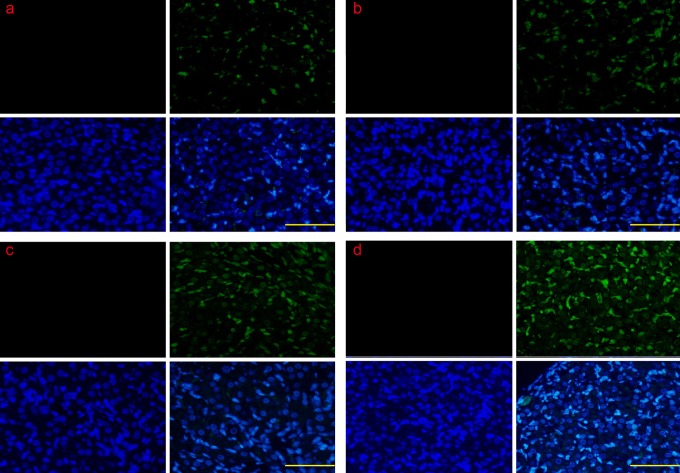
Figure 10.
The colocalization of ERS-associated apoptotic protein CHOP and caspase 12 by CLSM. The CHOP and caspase 12 protein were examined in early apoptosis and late apoptosis. The CHOP and caspase 12 proteins were translocated from the cytoplasm into the nucleus. A: control group; B: PGF2α-treated group; C: TUDCA-treated group; E: PGF2α+ TUDCA-treated group. Scale bar: a, b, c, and d: 100 μm. ERS indicates endoplasmic reticulum; CHOP, CCAAT/enhancer-binding protein homologous protein; CLSM, confocal laser scanning microscopy; PGF2α, prostaglandin F2α; TUDCA, tauroursodeoxycholic acid.
Figure 11.
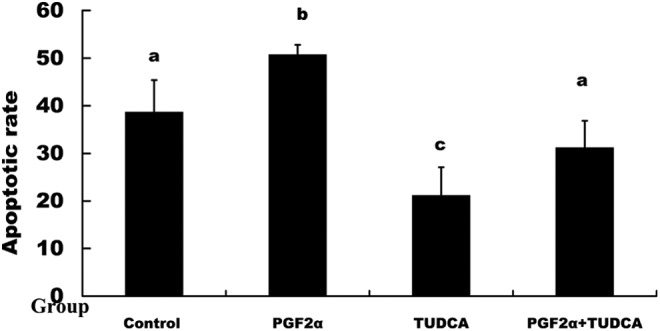
The analysis of luteal cell apoptosis by flow cytometry. The apoptotic rate of the group treated with PGF2α was significantly higher than that of the other groups. The apoptotic rate of the group treated with TUDCA was significantly lower than that of the other groups. The different letters indicate significant differences, and the same letters indicate no difference. PGF2α indicates prostaglandin F2α; TUDCA, tauroursodeoxycholic acid.
Discussion
Apoptosis of the luteal cells has been examined during spontaneous and induced CL regression in several animal species,6–12 and the death receptor- and mitochondria-mediated apoptotic pathways are involved in the apoptosis of luteal cells.13,15 A previous study suggested that hormonal-induced pseudopregnant rat ovaries were used as the model for regression of the CL, and a pseudopregnant CL could only be maintained for a certain number of days, after which spontaneous regression occurs.
Our results suggested that apoptosis of the luteal cells occurs in CL regression, and the number of apoptotic luteal cells was remarkably increased during the late-luteal stage. To study the effect of ERS on regression, the ERS marker molecule and apoptotic molecules were examined, and the ERS- and UPR-related molecules Grp78, CHOP, ATF6α, and caspase 12 were detected and changed during regression of CL. The levels were especially increased in the late-luteal stage of regression. The apoptotic execution molecule active caspase 3 was remarkably increased in the late-luteal stage of regression and the level peaked on day 20. In agreement with the messenger RNA levels, the expression levels of Grp78, CHOP, ATF6α, and caspase-12 proteins were found to be remarkably increased in the late-luteal stage of regression. The apoptotic execution molecule active caspase 3 was mainly detected in the late-luteal stage of regression and was localized in the nucleus. IRE1α, xbp1s, and eIF2α were not detected, and these results suggest that the apoptosis of luteal cells was mediated by ERS through the ATF6α-CHOP and caspase-12 pathways. Several studies have shown that the ATF6α-CHOP pathway existed in cellular apoptosis.23,43 Caspase 12 mediates ERS-specific apoptosis44 and is activated in the ERS-induced apoptosis.45 The apoptotic execution molecular caspase 3 is the downstream target of ERS-induced apoptosis.46 The mechanism of follicle atresia is the apoptosis of granulosa cells, and our previous study suggested that ERS is involved in follicle atresia and the apoptosis of granulosa cells.32,33 The regression of the CL and the apoptosis of luteal cells might be regulated by ERS. Indeed, our results further suggested by previous data and which study revealed that the ERS-mediated apoptotic pathway might be involved in the bovine CL regression during estrous cycle.16 Indeed, CHOP was detected in bovine and rat’s CL regression; in addition, the phospho-JNK signaling pathway was also detected in the bovine CL regression, combined with our result that caspase 12 is involved in rat’s CL regression. All results suggested that the ERS-mediated apoptosis pathway might be involved in the CL regression through CHOP, pJNK, and caspase-12 signaling pathways.
To confirm this suggestion, apoptosis of luteal cells was induced in vitro, and the ERS molecules were examined. The expression levels of the ERS-related proapoptotic molecules CHOP and caspase 12 were increased in the group with PGF2α and decreased in the group with TUDCA. The apoptotic execution marker active caspase 3 was increased in the group with PGF2α and decreased in the group with TUDCA. Apoptosis of the luteal cells was detected by flow cytometry, and the apoptotic rate was significantly higher in the group with PGF2α than in the other groups and significantly lower in the group with TUDCA than in the other groups. CHOP and caspase 12 were translocated from the cytoplasm to the nucleus during the late stage of apoptosis. The translocation of CHOP and caspase 12 has been detected in several studies.47–49 These findings suggested that ERS is involved in the regression of the CL and the apoptosis of luteal cells. Type II programmed cell death autophagy is a novel cellular death pathway, and apoptosis is type I programmed cell death. A previous study reported that autophagy is involved in the regression of CL and related to the apoptosis of luteal cells.3 A previous study suggested that the increased expression of bax promotes cellular apoptosis and bcl2 inhibits cellular apoptosis, and the ratio of bax to bcl2 is a critical determinant of apoptotic cell death.50,51 In accord with the results from previous studies,3 the expression levels of bax and bcl2 were examined, and the results suggested that bax increased but bcl2 decreased with increased apoptosis during induced regression of the CL. The apoptosis of regression of the CL is regulated by complicated pathways, and these pathways might be interlinked with each other. Endoplasmic reticulum stress is involved in the regression of the CL through the CHOP pathway and caspase 12.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article: This work was supported by Ningxia natural science funds (NZ13270).
Reference
- 1. Bowen-Shauver JM, Gibori G. The corpus luteum of pregnancy. In: Leung PCK, Adashi EY. (eds.), The Ovary. San Diego: Elseiver Inc., Academic Press; 2004:201–230. [Google Scholar]
- 2. Stouffer RL. The function and regulation of cell populations comprising the corpus luteum during the ovarian cycle. In: Leung PCK, Adashi EY. (eds.), The Ovary. San Diego: Elseiver Inc., Academic Press; 2004:169–184. [Google Scholar]
- 3. Choi J, Jo M, Lee E, Choi D. The role of autophagy in corpus luteum regression in the rat. Biol Reprod. 2011;85(3):465–472. [DOI] [PubMed] [Google Scholar]
- 4. Davis John S, Rueda Bo R. Recent advancements in corpus luteum development, function, maintenance and regression: Forum introduction. Reprod Biol Endocrinol. 2003;1(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stocco C, Telleria C, Gibori G. The molecular control of corpus luteum formation, function, and regression. Endocr Rev. 2007;28(1):117–149. [DOI] [PubMed] [Google Scholar]
- 6. Juengel JL, Garverick HA, Johnson AL, Youngquist RS, Smith MF. Apoptosis during luteal regression in cattle. Endocrinology. 1993;132(1):249–254. [DOI] [PubMed] [Google Scholar]
- 7. Rueda BR, Tilly KI, Botros IW, et al. Increased bax and interleukin-1beta-converting enzyme messenger ribonucleic acid levels coincide with apoptosis in the bovine corpus luteum during structural regression. Biol Reprod. 1997;56(1):186–193. [DOI] [PubMed] [Google Scholar]
- 8. Bowen JM, Keyes PL, Warren JSm, Townson DH. Prolactin-induced regression of the rat corpus luteum: expression of monocyte chemoattractant protein-1 and invasion of macrophages. Biol Reprod. 1996;54(5):1120–1127. [DOI] [PubMed] [Google Scholar]
- 9. Gaytan F, Morales C, Bellido C, et al. Progesterone on an oestrogen background enhances prolactin-induced apoptosis in regressing corpora lutea in the cyclic rat: possible involvement of luteal endothelial cell progesterone receptors. J Endocrinol. 2000;165(3):715–724. [DOI] [PubMed] [Google Scholar]
- 10. Telleria CM, Goyeneche AA, Cavicchia JC, Stati AO, Deis RP. Apoptosis induced by antigestagen RU486 in rat corpus luteum of pregnancy. Endocrine. 2001;15(2):147–155. [DOI] [PubMed] [Google Scholar]
- 11. Rueda BR, Wegner JA, Marion SL, Wahlen DD, Hoyer PB. Internucleosomal DNA fragmentation in ovine luteal tissue associated with luteolysis: in vivo and in vitro analyses. Biol Reprod. 1995;52(2):305–312. [DOI] [PubMed] [Google Scholar]
- 12. Shikone T, Yamoto M, Kokawa K, Yamashita K, Nishimori K, Nakano R. Apoptosis of human corpora lutea during cyclic luteal regression and early pregnancy. J Clin Endocrinol Metab. 1996;81(6):2376–2380. [DOI] [PubMed] [Google Scholar]
- 13. Quirk SM, Harman RM, Huber SC, Cowan RG. Responsiveness of mouse corpora luteal cells to Fas antigen (CD95)-mediated apoptosis. Biol Reprod. 2000;63(1):49–56. [DOI] [PubMed] [Google Scholar]
- 14. Carambula SF, Pru JK, Lynch MP, et al. Prostaglandin F2alpha- and FAS-activating antibody induced regression of the corpus luteum involves caspase-8 and is defective in caspase-3 deficient mice. Reprod Biol Endocrinol. 2003;1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dauffenbach LM, Khan SM, Yeh J. Corpus luteum regression in the rat in vivo and in vitro studies of apoptotic mechanisms. J Med. 2003;34(1–6):87–100. [PubMed] [Google Scholar]
- 16. Park HJ, Park SJ, Koo DB, et al. Unfolding protein response signaling is involved in development, maintenance, and regression of the corpus luteum during the bovine estrous cycle. Biochem Biophys Res Commun. 2013;441(2):344–350. [DOI] [PubMed] [Google Scholar]
- 17. Kogure K, Nakamura K, Ikeda S, et al. Glucose-regulated protein, 78-kilodalton is a modulator of luteinizing hormone receptor expression in luteinizing granulosa cells in rats. Biol Reprod. 2013;88(1):8. [DOI] [PubMed] [Google Scholar]
- 18. Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35(4):373–381. [DOI] [PubMed] [Google Scholar]
- 19. Kim R, Emi M, Tanabe K, Murakami S. Role of the unfolded protein response in cell death. Apoptosis. 2006;11(1):5–13. [DOI] [PubMed] [Google Scholar]
- 20. Rasheva VI, Domingos PM. Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis. 2009;14(8):996–1007. [DOI] [PubMed] [Google Scholar]
- 21. Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol. 2011;23(2):143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsutsumi S, Gotoh T, Tomisato W, et al. Endoplasmic reticulum stress response is involved in nonsteroidal anti-inflammatory drug-induced apoptosis. Cell Death Differ. 2004;11(9):1009–1016. [DOI] [PubMed] [Google Scholar]
- 23. Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–1030. [DOI] [PubMed] [Google Scholar]
- 24. Zinszner H, Kuroda M, Wang X, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12(7):982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beuers U. Drug insight: Mechanisms and sites of action of ursodeoxycholic acid in cholestasis. Nat Clin Pract Gastroenterol Hepatol. 2006;3(6):318–328. [DOI] [PubMed] [Google Scholar]
- 26. Ozcan U, Yilmaz E, Ozcan L, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang JY, Diao YF, Kim HR, Jin DI. Inhibition of endoplasmic reticulum stress improves mouse embryo development. PLoS One. 2012;7(7):e40433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J, Zhu G, Wang X, Xu B, Hu L. Apoptosis and expression of protein TRAIL in granulosa cells of rats with polycystic ovarian syndrome. J Huazhong Univ Sci Technolog Med Sci. 2007;27(3):311–314. [DOI] [PubMed] [Google Scholar]
- 29. Kim JS, Song BS, Lee KS, et al. Tauroursodeoxycholic Acid Enhances the Pre-implantation Embryo Development by Reducing Apoptosis in Pigs. Reprod Domest Anim. 2012;47(5):791–798. [DOI] [PubMed] [Google Scholar]
- 30. Abraham T, Pin CL, Watson AJ. Embryo collection induces transient activation of XBP1 arm of the ER stress response while embryo vitrification does not. Mol Hum Reprod. 2012;18(5):229–242. [DOI] [PubMed] [Google Scholar]
- 31. Hao L, Vassena R, Wu G, et al. The unfolded protein response contributes to preimplantation mouse embryo death in the DDK syndrome. Biol Reprod. 2009;80(5):944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin P, Yang Y, Li X, et al. Endoplasmic reticulum stress is involved in granulosa cell apoptosis during follicular atresia in goat ovaries. Mol Reprod Dev. 2012;79(6):423–432. [DOI] [PubMed] [Google Scholar]
- 33. Yang Y, Lin P, Chen F, et al. Luman recruiting factor regulates endoplasmic reticulum stress in mouse ovarian granulosa cell apoptosis. Theriogenology. 2013;79(4):633–639. [DOI] [PubMed] [Google Scholar]
- 34. Simmons D, Kennedy T. Induction of glucose-regulated protein 78 in rat uterine glandular epithelium during uterine sensitization for the decidual cell reaction. Biol Reprod. 2000;62(5):1168–1176. [DOI] [PubMed] [Google Scholar]
- 35. Liu AX, He WH, Yin LJ, et al. Sustained endoplasmic reticulum stress as a cofactor of oxidative stress in decidual cells from patients with early pregnancy loss. J Clin Endocrinol Metab. 2011;96(3):E493–E497. [DOI] [PubMed] [Google Scholar]
- 36. Iwawaki T, Akai R, Yamanaka S, et al. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci U S A. 2009;106(39):16657–16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lian IA, Løset M, Mundal SB, et al. Increased endoplasmic reticulum stress in decidual tissue from pregnancies complicated by fetal growth restriction with and without pre-eclampsia. Placenta. 2011;32(11):823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yung HW, Calabrese S, Hynx D, et al. Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction. Am J Pathol. 2008;173(2):451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Z, Wang H, Xu ZM, et al. Cadmium-induced teratogenicity: association with ROS-mediated endoplasmic reticulum stress in placenta. Toxicol Appl Pharmacol. 2012;259(2):236–247. [DOI] [PubMed] [Google Scholar]
- 40. Burton G, Yung HW, Cindrova-Davies T, Charnock-Jones DS. Placental endoplasmic reticulum stress and oxidative stress in the pathophysiology of unexplained intrauterine growth restriction and early onset preeclampsia. Placenta. 2009;30(suppl A):S43–S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Løset M, Mundal SB, Johnson MP, et al. A transcriptional profile of the decidua in preeclampsia. Am J Obstet Gynecol. 2011;204(1):84.e1–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kizuka F, Tokuda N, Takagi K, et al. Involvement of bone marrow-derived vascular progenitor cells in neovascularization during formation of the corpus luteum in mice. Biol Reprod. 2012;87(3):55. [DOI] [PubMed] [Google Scholar]
- 43. Araki E, Oyadomari S, Mori M. Endoplasmic reticulum stress and diabetes mellitus. Intern Med. 2003;42(1):7–14. [DOI] [PubMed] [Google Scholar]
- 44. Nakagawa T, Zhu H, Morishima N, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403(6765):98–103. [DOI] [PubMed] [Google Scholar]
- 45. Shibata M, Hattori H, Sasaki T, Gotoh J, Hamada J, Fukuuchi Y. Activation of caspase-12 by endoplasmic reticulum stress induced by transient middle cerebral artery occlusion in mice. Neuroscience. 2003;118(2):491–499. [DOI] [PubMed] [Google Scholar]
- 46. Hitomi J, Katayama T, Taniguchi M, Honda A, Imaizumi K, Tohyama M. Apoptosis induced by endoplasmic reticulum stress depends on activation of caspase-3 via caspase-12. Neurosci Lett. 2004;357(2):127–130. [DOI] [PubMed] [Google Scholar]
- 47. Shiraishi H, Okamoto H, Yoshimura A, Yoshida H. ER stress-induced apoptosis and caspase-12 activation occurs downstream of mitochondrial apoptosis involving Apaf-1. J Cell Sci. 2006;119(pt 19):3958–3966. [DOI] [PubMed] [Google Scholar]
- 48. Sokka AL, Putkonen N, Mudo G, et al. Endoplasmic reticulum stress inhibition protects against excitotoxic neuronal injury in the rat brain. J Neurosci. 2007;27(4):901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu X, Li Y, Wang W, et al. 3β-Hydroxysteroid-Δ 24 Reductase (DHCR24) Protects Neuronal Cells from Apoptotic Cell Death Induced by Endoplasmic Reticulum (ER) Stress. PLoS One. 2014;9(1):e86753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74(4):609–619. [DOI] [PubMed] [Google Scholar]
- 51. Williams GT, Smith CA. Molecular regulation of apoptosis: genetic controls on cell death. Cell. 1993;74(5):777–779. [DOI] [PubMed] [Google Scholar]