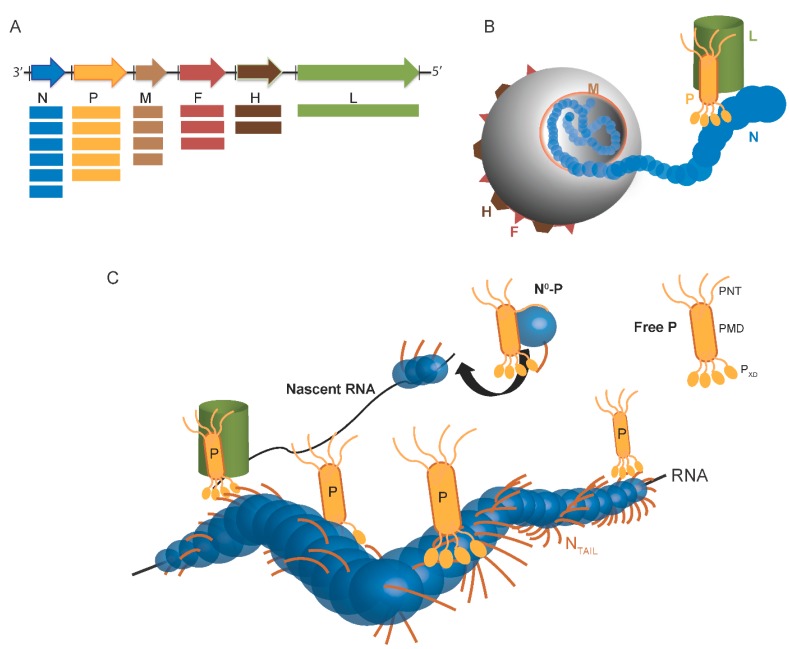
Figure 1.
Scheme of Paramyxovirinae genome and viral particule. (A) Schematic representation of the genome of Paramyxovirinae. The negative-sense genomic RNA is presented in the 3′ to 5′ orientation. The open reading frames are represented by colored arrows and encode the nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), attachment protein (H) and large polymerase protein (L). Vertical lines represent gene start and stop signals. Below the genome, shown is a schematic representation of the expression gradient of the encoded proteins as a result of the stop and re-initiation mechanism of the polymerase during transcription [2]; (B) Schematic illustration of the virion. The viral membrane is decorated by the F and H glycoproteins and M is located beneath the membrane. N is bound to genomic RNA and together with P and L forms the viral replication unit; (C) Schematic illustration of the Paramyxoviridae replicative complex. The RNA is represented as a solid black line. The neo-synthetized RNA is shown already partially encapsidated by N. The N and P intrinsically disordered regions are symbolized by lines. The extended conformation of the disordered regions is thought to allow the formation of a tripartite complex between N°, P and L required for nucleocapsid assembly. The P/L complex forms the RNA-dependent RNA polymerase (RdRp) complex that cartwheels onto the nucleocapsid complex via the X domain of P (PXD). P is shown as a tetramer to reflect the prevalence of this oligomeric state in paramyxoviral P proteins.