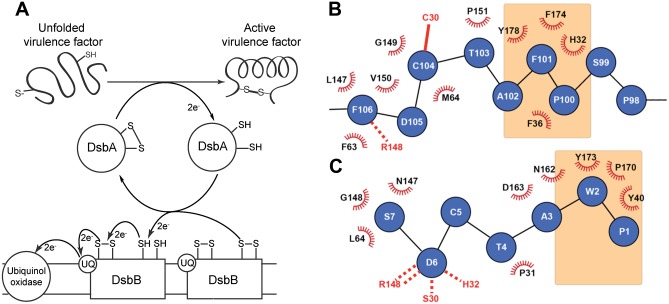
Fig 1. The DsbA-DsbB interaction.
A. Schematic showing the proposed mechanism of oxidative folding in the periplasm of Gram-negative bacteria. DsbA catalyses the formation of a disulfide bond in a protein substrate, then interacts with DsbB to which it transfers electrons so that DsbA is regenerated into its active oxidized state. The electrons are subsequently transferred from DsbB to ubiquinone (UQ) and ultimately to the respiratory complex. B. The binding interface between EcDsbA (black and red) and EcDsbB loop P2 (blue) derived from the crystal structure of the EcDsbAC33A:EcDsbBC130S complex [37]. The EcDsbA hydrophobic groove residues are highlighted in orange shading, the intermolecular disulfide bond is shown as a solid red line and the hydrogen bond with the cisPro loop is shown as a dashed red line. C. The binding interface between PmDsbA (black and red) and the peptide PWATCDS (blue) from the crystal structure of the complex [38]. In this complex there is no disulfide bond as the active site cysteine of PmDsbA was mutated to Ser (S30). The peptide Cys5 residue points away from the binding interface. Residues W2 and P1 of the peptide both interact with the hydrophobic groove (in orange) and these interactions were used as the target for this peptidomimetic design.