Fig 5.
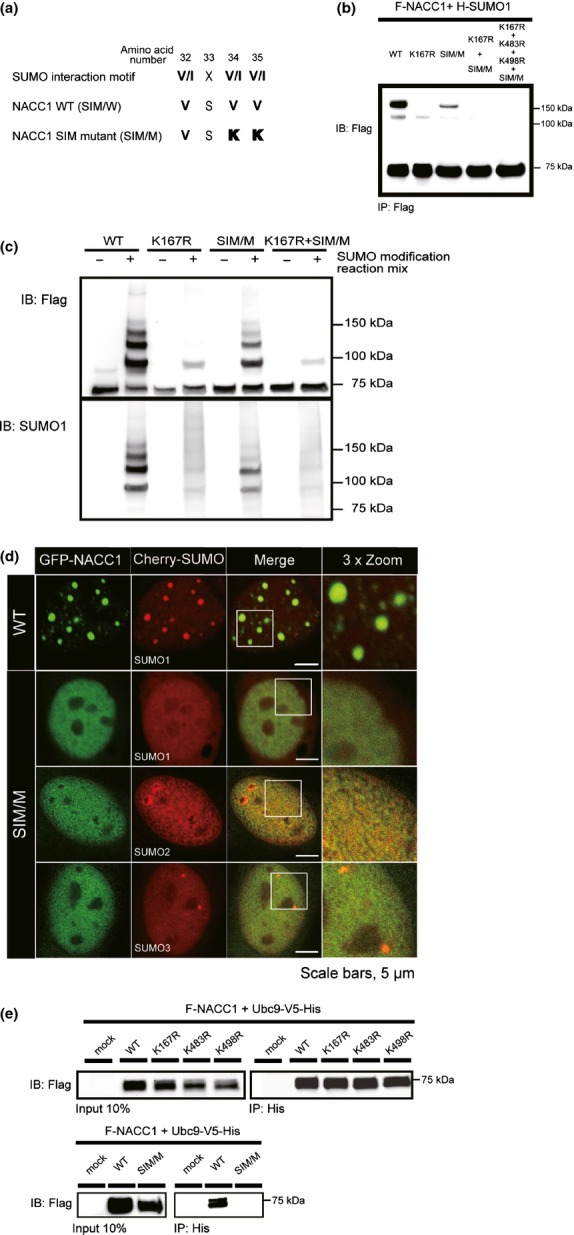
Disruption of the SIM sequence of nucleus accumbens associated 1 (NACC1) and the interaction with NACC1 and UBC9. (a) SIM consensus motif aligned with the NACC1-WT and NACC1-SIM mutant sequence (SIM/M). (b) HeLa cells were transfected with a plasmid encoding either F-NACC1-WT or F-NACC1-SIM/M, together with H-SUMO1. Slow-migrating bands were observed with SIM/M, but their density was reduced in comparison with those of the WT. A weak slow-migrating band was found in K167R but not in K167R + SIM/M or K167R + K483R + K498R + SIM/M. (c) Analysis of SUMO modification by reconstituted in vitro SUMOylation reactions. Slow-migrating bands due to SUMO1 were observed in WT and SIM/M, although the density of the migrating bands was weaker in SIM/M than in WT and was further reduced in K167R and K167R + SIM/M. (d) Confocal immunofluorescence of GFP-NACC1-WT or -SIM/M mutants and Cherry-SUMO1/2/3 fusion proteins (NACC1, green; SUMO1/SUMO2/SUMO3, red). Both fusion proteins colocalized in WT with a discrete nuclear body pattern. Complete disruption of the nuclear body pattern was observed when the SIM/M construct was used. Bar, 5 μm. (e) Cotransfection and immunoprecipitation analyses for the binding between UBC9 and NACC1. HeLa cells were cotransfected with plasmids encoding various F-NACC1-WT, -K167R, -K483R, -K498 or -SIM/M and UBC9-V5/His vectors. The interaction between NACC1 and UBC9 was suppressed only in the case of Flag-NACC1 SIM/M.
