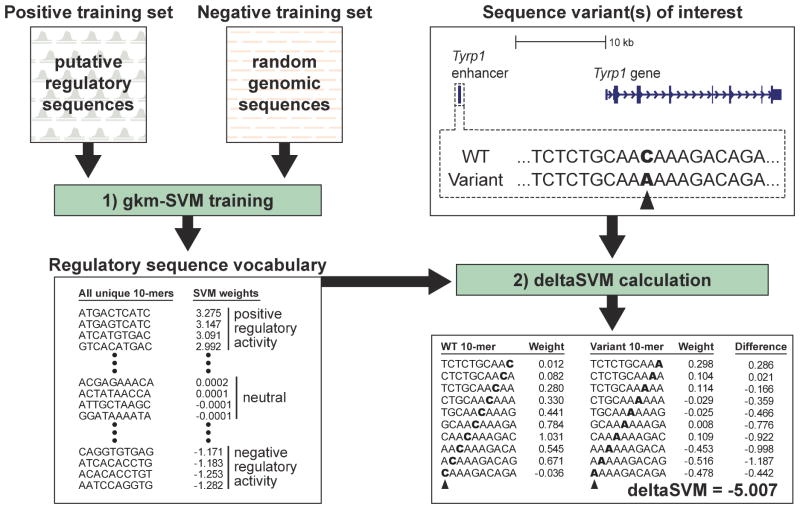
Figure 1. Overview of our deltaSVM method.
[left] The first step in calculating deltaSVM is to train a gkm-SVM classifier using a positive training set of putative regulatory sequences (identified by DNase I hypersensitivity, for example) and a negative training set of matched negative control sequences. The gkm-SVM generates a regulatory sequence vocabulary – a weighted list of all possible 10-mers, where each 10-mer receives an SVM weight that quantifies its contribution to the prediction of regulatory function. [right] After training, this regulatory sequence vocabulary can be used to score the predicted impact of any sequence variant on regulatory activity, as shown here for a single nucleotide substitution in a melanocyte enhancer of the Tyrp1 enhancer.