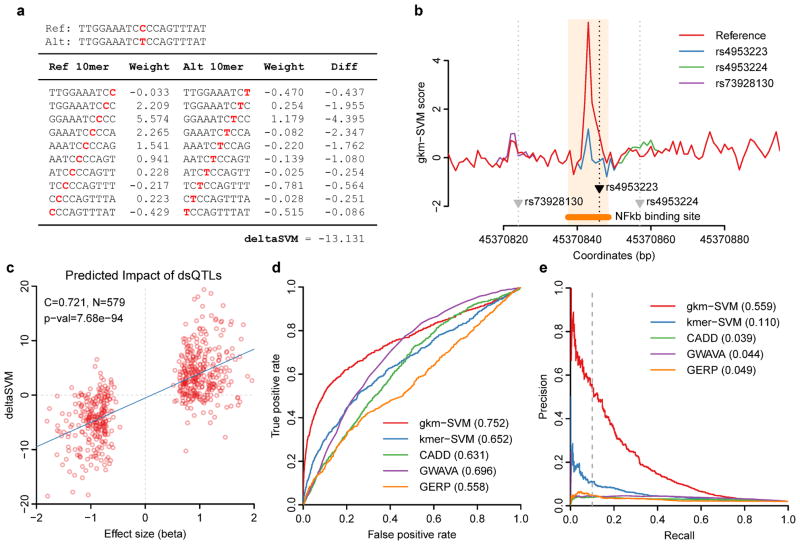
Figure 2. deltaSVM can accurately predict SNPs associated with DNaseI Hypersensitivity.
(a) An example of a deltaSVM calculation using a known dsQTL SNP (rs4953223). (b) 10-mer gkm-SVM scores across the dsQTL locus containing rs4953223 are shown. Only the functional SNP produces dramatic changes in gkm-SVM scores. (c) Effect sizes of dsQTL SNPs from Ref. 13 are well correlated with their deltaSVM scores. (d–e) deltaSVM predicts dsQTLs with far greater accuracy than existing methods. Discriminative powers are compared between various methods using 50x larger control SNP set. (d) ROC curve. (e) Precision-Recall curve.