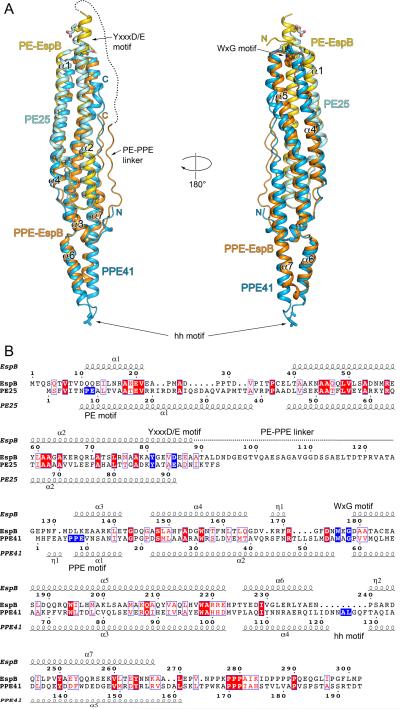
Fig. 3.
Comparison of EspB and PE25–PPE41 structures.
A. Structural superposition of EspB7-278 (PDB: 4XXX) and PE25–PPE41 dimer (PDB: 4KXR). PE domain of EspB is in yellow; PPE domain of EspB is in orange; PE25 is in cyan; PPE41 is in light blue. The disordered region of PE–PPE linker of EspB is indicated by a dashed line. The secondary structure elements of EspB are labeled. The hh motif residues of PPE41, and the YxxxD/E motif residues of EspB and PE25 are shown in stick representation. Structures were superimposed using Secondary Structure Matching (SSM) (Krissinel and Henrick, 2004) as implemented in Coot (Emsley et al., 2010).
B. Structure-based sequence alignment of EspB and PE25–PPE41 based on superposition in (A). Secondary structure elements of EspB7-278 and PE25–PPE41 are displayed above and below sequences, respectively. The characteristic sequence motifs of EspB and PE25–PPE41 are highlighted in blue. The alignment figure was generated using the ESPript 3.0 server (http://espript.ibcp.fr/ESPript/ESPript/) (Robert and Gouet, 2014).