FIGURE 5.
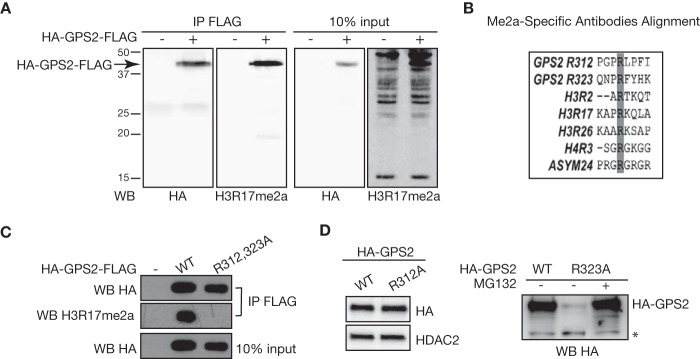
Methylation of GPS2 in vivo onArg-R312 and Arg-323. A, immunoprecipitation (IP) for FLAG followed by Western blotting with the pan-arginine methylation antibody H3R17me2a confirms in vivo methylation of HA-GPS2-FLAG. Western blotting (WB) for HA provided a positive control for the efficiency of immunoprecipitation. B, alignment of the motifs around the GPS2 putative methylation sites Arg-312 and Arg-323 with the arginine methylation sites recognized by the antibodies tested in A. C, confirmation of in vivo methylation of GPS2 on Arg-312 and Arg-323 in 293T cells transfected with FLAG-eYFP, HA-GPS2WT-FLAG, or the HA-GPS2R312/323K-FLAG double mutant. Immunoprecipitation was followed by Western blotting with the H3R17me2a antibody as in A. D, Western blotting for HA-GPS2 in 293T nuclear extracts showed that mutation of arginine 323 alone (GPS2-R323A) is sufficient to promote GPS2 degradation in a proteasome-dependent manner (right panel), whereas mutating R312A does not affect GPS2 protein expression (left panel). Blotting for HDAC2 provided a loading control. The asterisk indicates a nonspecific band.
