FIGURE 1.
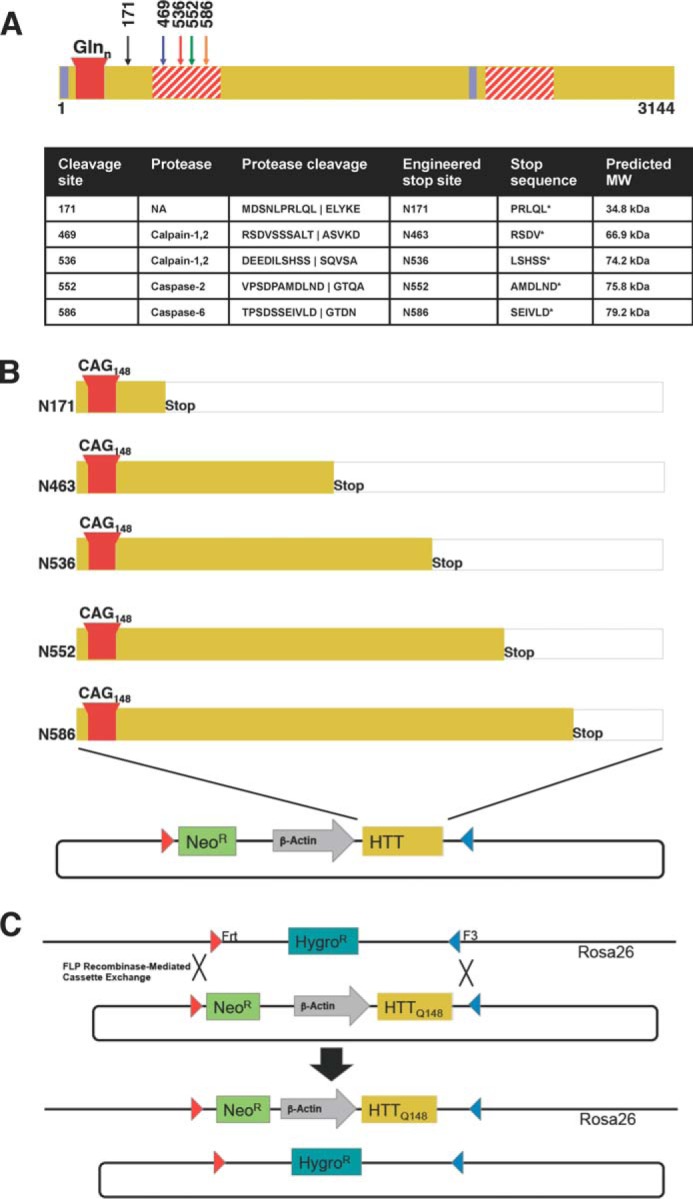
Schematic of proteolysis of HTT and the generation of Rosa-stop HD transgenic mice. A, schematic and table summary of the proteolysis events occurring in the HTT protein. Only the cleavage sites for fragments investigated in this paper are shown. The proteolytic susceptibility domains (red striped), nuclear export signals (blue), and polyQ stretch (red) are noted. HTT is cleaved by calpains-1 and -2 at amino acids 469 and 536, by caspase-2 at amino acid 552, and by caspase-6 at amino acid 586. B, design of HTT N-terminal fragment transgenic HD mouse models. mHTT N-terminal cleavage sites were modeled by engineering of a stop codon after the sites indicated, and these sites result in truncated protein fragments that are identical to caspase or calpain cleavage products with the exception of N463, which mimics cleavage site at an amino acid just before amino acid 469. Individual N-terminal fragment constructs in a donor vector flanked by Frt and F3 sites and engineered for recombinase-mediated cassette exchange in C57BL/6-Gt(ROSA)26Sortm596Arte mESCs are shown. C, schematic of the Flp recombinase-mediated cassette exchange in mESCs. Hygromycin resistant mESCs were transfected with recombinase and donor and selected for resistance to neomycin. Resulting clones were verified and used to generate chimeric mice with BALB/c embryos. Chimerism was assessed by coat color, and highly chimeric males were bred to C57BL/6 female mice to generate founders. Predicted average molecular masses of the fragments, calculated in the ExPASy Compute pI/Mw tool (81) are as follows: N171 Q145, 34.8 kDa; N463 Q145, 66.9 kDa; N536 Q145, 74.2 kDa; N552 Q145, 75.8 kDa; N586 Q145, 79.2 kDa. The fragments run higher than their calculated molecular masses as the expanded polyQ region reduces mobility in SDS-PAGE (82).
