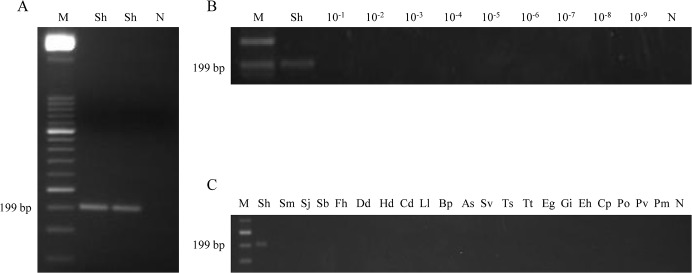
Fig 2. PCR verification, detection limit and specificity using outer primers F3 and B3.
(A) PCR verification of expected 199 bp target length amplicon. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lane Sh, S. haematobium DNA (1 ng); lane N, negative control (no DNA template). (B) Detection limit of PCR. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lane Sh: S. haematobium DNA (1 ng); lanes 10−1–10−9: 10-fold serially dilutions of S. haematobium DNA; lane N, negative control (no DNA template). (C) Specificity of PCR. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sh, Sm, Sj, Sb, Fh, Dd, Hd, Cd, Ll, Bp, As, Sv, Ts, Tt, Eg, Gi, Eh, Cp, Po, Pv, Pm, S. haematobium, S. mansoni, S. japonicum, S. bovis, Fasciola hepatica, Dicrocoelium dendriticum, Hymenolepis diminuta, Calicophoron daubneyi, Loa loa, Brugia pahangi, Anisakis simplex, Strongyloides venezuelensis, Trichinella spiralis, Taenia taeniformis, Echinococcus granulosus, Giardia intestinalis, Entamoeba histolytica, Cryptosporidium parvum, Plasmodium ovale, P. vivax and P. malariae DNA samples (1 ng/each), respectively; lane N, negative control (no DNA template).