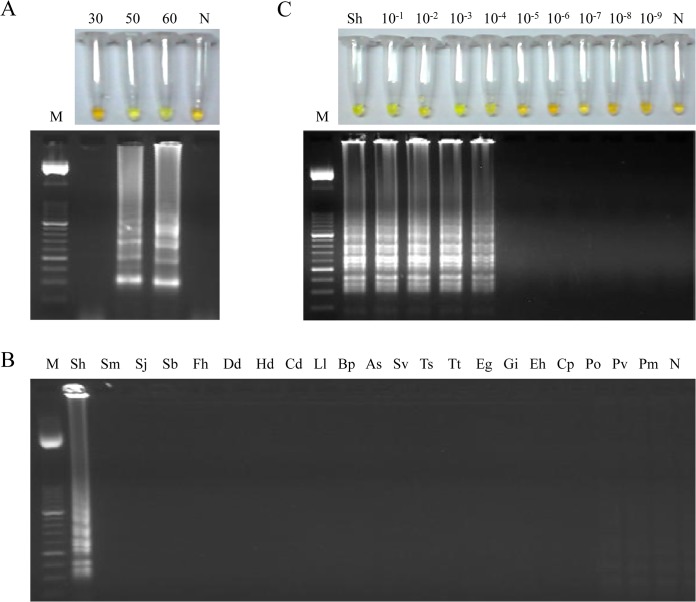
Fig 3. Setting up LAMP assay.
(A) LAMP amplification results obtained at different incubation times (30, 50 and 60 min) tested in a heating block by the addition of SYBR Green I (up) or by visualization on agarose gel (down). Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes 30, 50, 60, amplification results of S. haematobium DNA (1 ng) for 30, 50 and 60 minutes of incubation time, respectively. (B) Specificity of the LAMP assay for S. haematobium. A ladder of multiple bands of different sizes could be only observed in S. haematobium DNA sample. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sh, Sm, Sj, Sb, Fh, Dd, Hd, Cd, Ll, Bp, As, Sv, Ts, Tt, Eg, Gi, Eh, Cp, Po, Pv and Pm, S. haematobium, S. mansoni, S. japonicum, S. bovis, Fasciola hepatica, Dicrocoelium dendriticum, Hymenolepis diminuta, Calicophoron daubneyi, Loa loa, Brugia pahangi, Anisakis simplex, Strongyloides venezuelensis, Trichinella spiralis, Taenia taeniformis, Echinococcus granulosus, Giardia intestinalis, Entamoeba histolytica, Cryptosporidium parvum, Plasmodium ovale, P. vivax and P. malariae DNA samples (1 ng/each), respectively; lane N, negative control (no DNA template). (C) Sensitivity assessment performed with LAMP at 63°C for 50 min using serial dilutions of S. haematobium genomic DNA. Lane M: 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sh: genomic DNA from S. haematobium (1 ng); lanes 10−1–10−9: 10-fold serially dilutions; lane N: negative controls (no DNA template).