Abstract
Background
Schistosoma japonicum causes major public health problems in China and the Philippines; this parasite, which is transmitted by freshwater snails of the species Oncomelania hupensis, causes the disease intestinal schistosomiasis in humans and cattle. Researchers working on Schistosoma in Africa have described the relationship between the parasites and their snail intermediate hosts as coevolved or even as an evolutionary arms race. In the present study this hypothesis of coevolution is evaluated for S. japonicum and O. hupensis. The origins and radiation of the snails and the parasite across China, and the taxonomic validity of the sub-species of O. hupensis, are also assessed.
Methodology/Principal Findings
The findings provide no evidence for coevolution between S. japonicum and O. hupensis, and the phylogeographical analysis suggests a heterochronous radiation of the parasites and snails in response to different palaeogeographical and climatic triggers. The results are consistent with a hypothesis of East to West colonisation of China by Oncomelania with a re-invasion of Japan by O. hupensis from China. The Taiwan population of S. japonicum appears to be recently established in comparison with mainland Chinese populations.
Conclusions/Significance
The snail and parasite populations of the western mountain region of China (Yunnan and Sichuan) appear to have been isolated from Southeast Asian populations since the Pleistocene; this has implications for road and rail links being constructed in the region, which will breach biogeographical barriers between China and Southeast Asia. The results also have implications for the spread of S. japonicum. In the absence of coevolution, the parasite may more readily colonise new snail populations to which it is not locally adapted, or even new intermediate host species; this can facilitate its dispersal into new areas. Additional work is required to assess further the risk of spread of S. japonicum.
Author Summary
Schistosomiasis is a disease caused by a parasitic worm transmitted to humans by certain species of freshwater snails. In spite of several decades of intensive coordinated control schistosomiasis still infects around 1 million people in China. In order to understand the potential for spread of the disease into new areas and new snail species, it is helpful to know if the snails and parasites in China are coevolved; this means that evolutionary divergence in one group (the snails) is matched by a corresponding divergence in the other (the parasites), which is what would be expected if the two groups are locked in an evolutionary arms race. DNA-sequence data were collected for snails and parasites from the same localities. The findings indicated that coevolution was unlikely to have occurred. The implications of this are that host-switching or acquisition is more likely than previously thought. Consequently, there is a greater potential for spread of the parasite into new areas. The role of mountain barriers in confining schistosomiasis to certain regions was highlighted; this is important in view of the current plans to breach these barriers by road and rail construction that will link China and Southeast Asia.
Introduction
Background
In China schistosomiasis in humans is caused by infection with the parasitic blood-fluke Schistosoma japonicum (Trematoda: Digenea). Schistosomiasis causes major public health problems in China and the Philippines[1]. Despite over 45 years of integrated control efforts, approximately one million people, and more than 1.7 million bovines and other mammals, are currently infected in mainland China[2]. The disease develops after direct penetration of the skin by free-swimming parasite larvae (cercariae) released from the snail intermediate host. Exposure to cercariae of S. japonicum occurs through contact with wet vegetation or walking through rice fields or other flooded areas inhabited by infective snails. In the case of S. japonicum, only sub-species of the snail Oncomelania hupensis (Gastropoda: Pomatiopsidae) are able to act as intermediate host[3]. The snails become infected by similarly mobile and penetrative larvae (miracidia) passed in the feces of definitive hosts, which include a wide variety of mammals (up to 28 genera and 7 orders [1,4,5]).
Much of the transmission in mainland China occurs across the generally flat and marshy lake-land areas around the middle to lower Yangtze river, where Oncomelania hupensis is the snail intermediate host. In the highland areas of Southwest China (Sichuan and Yunnan) O. hupensis robertsoni is the intermediate host. Fewer people are infected in the highland areas; however, recrudescence is most marked in this region and in Sichuan between 1989 and 2004, the disease re-emerged in 8 counties (prevalence: 1.4% to 18.3% in humans, and up to 22.3% in cattle and 0.16% in snails)[6]. A third subspecies is found in China, O. hupensis tangi, which is endemic to hilly or coastal areas of Fujian, Guangdong and Guangxi provinces, although S. japonicum transmission appears to have been interrupted in these areas. In the Philippines, a further sub-species is responsible for transmission, namely O. hupensis quadrasi, and in Sulawesi O. hupensis lindoensis is responsible. Other subspecies exist in Taiwan and in Japan (where the only other species of Oncomelania, O. minima, is also found) but these are not, or are no longer, relevant to human health[7].
Clearly, S. japonicum continues to pose a serious public health problem and inter-disciplinary research is required to understand the patterns of transmission and persistence of the disease. Phylogeographical studies can shed light on the problem, particularly in determining where the disease comes from, explaining current distributions of the intermediate host and predicting future epidemiology. Comparative phylogenetics can help detect patterns of host-parasite coevolution and indicate any potential for regional adaptation. Despite the potential of such approaches, relatively little work has been done in this area for S. japonicum. The present study was therefore performed in order to apply current phylodynamic techniques to the estimation of sources and tracts of dispersal for this parasite and to test for the signatures of host-parasite coevolution during the evolution of S. japonicum and its intermediate hosts (the latter necessarily having the defining influence on the distribution of the parasite). Strictly, the techniques used here test for phylogenetic congruence rather than directly for coevolution. The absence of phylogenetic congruence would make long-term coevolution unlikely; thus providing an indirect test for the latter. The study aimed to utilise the largest and most geographically extensive data set available (Fig 1).
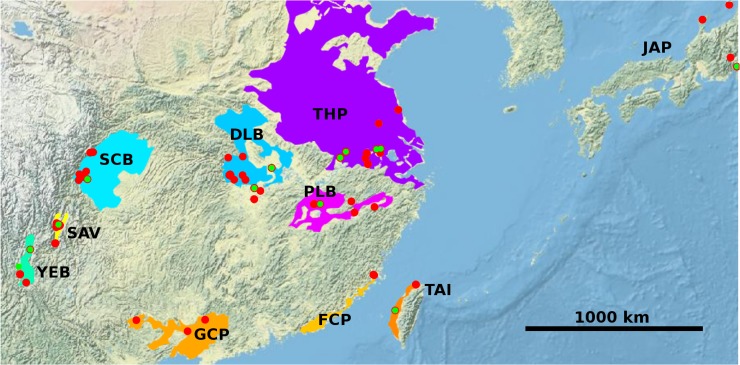
Fig 1. The locations of the sample sites in China and Japan.
The coloured spots indicate areas from which snail and worm populations were sampled (red and green respectively). Most of the red spots lie on top of a green spot, indicating that the snails and the worms were sampled from the same locality. The coloured regions define biogeographical areas, within which there are no significant barriers to snail dispersal (e.g., no high mountains). Philippine samples (worms and snails) were included, but omitted from the map to increase resolution. Abbreviations are listed in Table 1. Plotted using the OpenStreetMap package in R.
A geographical strain complex has been revealed within Schistosoma japonicum using partial DNA sequences of mitochondrial genes [22,23]. In 2005 microsatellites were used to assess variation among S. japonicum populations from 8 geographical locations in 7 endemic provinces across mainland China. The populations were found to fall into two main clades, a Sichuan/Yunnan clade and a lake/marshland clade of the middle and lower Yangtze river drainage[24]. The finding was later corroborated[25] using partial mitochondrial gene sequences (cytb-nd4L-nd4, cox3, nad1 nad4, nad5 and 16S-12S), with the additional observation that the lake/marshland clade included a highly diverse lower Yangtze sub-clade[25,26]. Molecular phylogenetic studies of Oncomelania hupensis are relatively few in number, but a major study in 2009[19] suggested that O. hupensis populations across China fall into three main clades. These were a middle/lower Yangtze lake and a marshland clade (O. hupensis hupensis), a Sichuan/Yunnan mountain clade (O. h. robertsoni), a clade corresponding to populations of the hilly littoral areas of Fujian (O. h. tangi) and a clade of the karst region of Guangxi (O. h. hupensis or, according to the 2009 authors, O. h. guangxiensis). A more recent study used complete mitochondrial genome sequences to obtain mean genetic distance estimate of 12% for the protein coding genes between O. h. hupensis and O. h. robertsoni[13]. A study with similarly wide geographical coverage to that used here has been performed[27]; this employed DNA sequence variation in O. hupensis to indicate that isolation by distance was more significant in shaping genetic divergence than isolation by environment; however, this study which covered 29 localities, was population genetic (not comparative phylogenetic) and did not include the parasite.
Aims of the study
Although there have been past population phylogenetic studies of both Oncomelania and S. japonicum none has used as geographically diverse, taxon and character rich, data set as the present study. Also, this is the first quantitative comparison of the phylogenetic history of the snails and the parasites. Relatively few phylogenetic studies have considered the origins of these taxa, although Davis[28] proposed a Gondwanan origin for the Pomatiopsidae (including Oncomelania and proto-S. japonicum), with rafting to mainland Asia (via the Indian Craton after the break up of Gondwana) and colonisation of Southeast Asia and China (Tibet/Yunnan) via the northern-India-Myanmar, Brahmaputra-Irrawaddy river corridor in an West to East direction, around 18 Ma. In contrast, an East to West hypothesis has been proposed for the Chinese Pomatiopsidae taxa[29], with precursors of Oncomelania colonising originally from Australasia and via the Philippines, along island chains created by low sea levels and by tectonic movements (rafting). After reaching Japan, Proto-Oncomelania gives rise to Oncomelania hupensis in mainland China; the latter then recolonises Japan, the Philippines and Sulawesi (replacing antecedent forms). In a recent paper[30], the East to West hypothesis was tested and the radiation across China dated at 15–5 Ma (by molecular clock); however, the radiation of S. japonicum did not appear to be isochronous with that of the present day intermediate hosts[31,32]. Nevertheless, the history of host utilization in Schistosoma has been regarded as an evolutionary battle with snail defences[11,33–37], with the schistosome under significant pressure to evolve counter measures to snail immune responses, or to track snail divergence in an evolutionary arms race (the ‘Red Queen hypothesis’[38]).
Schistosomiasis researchers postulating coevolution have evidenced this by citing restriction of the parasites to certain snail lineages[11], high levels of genetic variation in naturally exposed snail populations[34], and evidence for selection in schistosomes exposed to snails previously selected for resistance[36]. The latter study[36] did use genetic (microsatellite) variation to demonstrate higher rates of parasite divergence in resistant laboratory lines of snails; however, this was over a very short time-scale and the resistant populations had not evolved under natural conditions. The study also used the Schistosoma mansoni:Biomphalaria glabrata system (which involves far higher infection rates than seen with S. japonicum–see Discussion). Consequently, further studies are needed into the role of coevolution in the evolutionary history of Schistosoma.
The present investigation was performed in view of the lack of studies on S. japonicum comparing the parasite and snail intermediate host evolutionary histories, the alternative hypotheses regarding the origins of these taxa and the colonisation of mainland China, and the recent availability of additional DNA sequence data (improving geographical coverage). It is important to address these questions because they can help explain the current distribution of the parasite within the range of the snails, which is relevant to snail control strategies and the potential for range expansion, and to help assess the likely impacts of environmental manipulation such as the South-to-North-Water-Transfer project in China and the construction of road links. The Greater Mekong Subregion (GMS) Chengdu-Kunming corridor and GMS North-South Corridor from Yunnan, through Laos and to Thailand (NSEC1), both involve tunnels and rapid links through the mountains of Sichuan and Yunnan to Laos[39]. Consequently, the work will not only improve our understanding of host-parasite coevolution, but also shed light on the impacts of development in the region.
Methods
Sampling
Three new samples of Oncomelania were taken and sequenced for this study; these were O. hupensis from Jiangshan in Jiangxi Province, China (cox1), and from Xinjin in Chengdu, Sichuan Province, China (cox1 and rrnL), and O. hupensis nosophora from Kanagawa Kiyokawa in Honshu, Japan (cox1 and rrnL)–these loci all belonging to the mitochondrial genome, with partial DNA sequences of each locus obtained using the popular Folmer[40] and Palumbi[41] primers, to allow comparisons with data from earlier studies. The three samples were identified on the basis of conchology, morphology and ecological habit (following[9]). The identifications were performed by YS and NH, two greatly experienced medical malacologists. The DNA sequencing followed procedures detailed elsewhere[30]. The remainder of the data were obtained from GenBank to give a total of 265 cox1 sequences and 70 rrnL sequences, apparently orthologous with the new sequence data above. Sequence data for Tricula hortensis from Sichuan China, also a pomatiopsid snail[42], were included for use as an outgroup. In addition to the data for Oncomelania, 14 complete cox1 and cox3 DNA sequences were obtained from the GenBank for samples from 14 Schistosoma japonicum transmission localities across China. A corresponding Schistosoma mekongi sequence was obtained as an outgroup taxon for the S. japonicum analyses. Full details of the data used and sampling range are given in Fig 1 and Tables 1, 2 and 3. The data set included all available relevant sequence data in the GenBank at the time of searching; however, some data (10 sequences) were excluded because of uncertain origin (e.g., DQ112269, DQ112270) and taxonomy (e.g., EU780224). The cox1 and cox3 loci were chosen following earlier work, which indicated that, for Schistosoma, cox3 was the locus of choice in terms of consistent phylogenetic signal and sufficient number of phylogenetically informative characters per site; this was followed by nad4, however, the present data showed a haplotype diversity <1 at this locus and cox1 was used because it gave high support values and “correct” topology in the same study[43]. Many of the localities listed in Tables 1 and 2 were not published either in GenBank or in the papers where the data were first presented. In these cases the localities were found by accessing field work reports cited in the paper presenting the sequence data (if present), matching published sample codes with associated accession numbers, reference to museum specimen accession numbers, contacting original researchers (or the local officials who accompanied them or those who arranged their collections) and referring to the personal observations of the present authors. In some cases, incomplete (e.g., to County level only) location data was given, or ambiguously transliterated place names, in these cases location records were completed and transliterations resolved. The locations in Tables 1 and 2 have been changed (where necessary) to use the closest place name (village etc) appearing on Google Maps; this is for the convenience of future authors.
Table 1. Malacological data used in the phylogeographical reconstructions–cox1 locus (CO1).
| GAC | Taxon | Locality | Coordinates | Ref |
|---|---|---|---|---|
| Fujian coastal plain (China)—FCP | ||||
| DQ212796 | Oncomelania hupensis tangi | Fuzhou, Fuqing, Donggang, Donghanzhen | 25.430833 119.605278 | [8] |
| JF284695 | Oncomelania hupensis tangi * | Fuzhou, Fuqing, Donggang, Donghanzhen, Nansha | 25.4049 119.6471 | unpub |
| Guangxi coastal plain (China)—GCP | ||||
| DQ112264 | Oncomelania hupensis hupensis | Xishanzhen, Guiping, Guigang (Yujiang river/Qian river) | 23.391 110.078 | [9] |
| DQ112265 | Oncomelania hupensis hupensis | Xishanzhen, Guiping, Guigang (Yujiang river/Qian river) | 23.391 110.078 | [9] |
| DQ112266 | Oncomelania hupensis hupensis | Xishanzhen, Guiping, Guigang (Yujiang river/Qian river) | 23.391 110.078 | [9] |
| GU367391 | Oncomelania hupensis | Nanning, Taoxuzhen, Dingwu (Haitan Dao coastal plain W) | 22.816667 109.116667 | [10] |
| JF284696 | Oncomelania hupensis | Nanning, Jing Xi Xian, Baise, Bameng Reservoir | 23.3655 106.2997 | unpub |
| Hunan / Hubei Dongting Lake Basin (China)—DLB | ||||
| AF253072 | Oncomelania hupensis hupensis | Hubei, Wuhan, Caidian, Beiban (Houguan Hou) | 30.569 114.017 | [8] |
| AF253073 | Oncomelania hupensis hupensis | Hubei, Wuhan, Caidian, Beiban (Houguan Hou) | 30.569 114.017 | [8] |
| AF254478 | Oncomelania hupensis hupensis | Hubei, Wuhan, Caidian, Beiban (Houguan Hou) | 30.569 114.017 | [11] |
| AF254479 | Oncomelania hupensis hupensis | Hubei, Wuhan, Caidian, Beiban (Houguan Hou) | 30.569 114.017 | [11] |
| AF254480-254489 | Oncomelania hupensis hupensis | Hunan, Yue Yang, Junshan, Tangang Cun (Yangtze River) | 29.617 113.067 | [11] |
| AF306572-306581 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Desheng Cun, Miao River (near Changhu) | 30.20139 111.765028 | [12] |
| AF306582 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Qichansi, Miao River (nr Changhu) | 30.25714 111.734641 | [12] |
| AF306583 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Qichansi, Miao River (nr Changhu) | 30.25714 111.734641 | [12] |
| AF306584 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Qichansi, Miao River (nr Changhu) | 30.25714 111.734641 | [12] |
| AF306585 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Qichansi, Miao River (nr Changhu) | 30.25714 111.734641 | [12] |
| AF306586 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Qichansi, Miao River (nr Changhu) | 30.25714 111.734641 | [12] |
| AF306587 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Qichansi, Miao River (nr Changhu) | 30.25714 111.734641 | [12] |
| AF306588-306597 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Guanyue Cun, Miao River (near Changhu) | 30.22888 111.692471 | [12] |
| AF306598 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Sunhe Supermarket Guanyue Cun | 30.229074 111.691602 | [12] |
| AF306599 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Sunhe Supermarket Guanyue Cun | 30.229074 111.691602 | [12] |
| AF306600 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Sunhe Supermarket Guanyue Cun | 30.229074 111.691602 | [12] |
| AF306601 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Sunhe Supermarket Guanyue Cun | 30.229074 111.691602 | [12] |
| AF306602 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Sunhe Supermarket Guanyue Cun | 30.229074 111.691602 | [12] |
| AF306603-306612 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Longtanhe Cun, Miao River (near Changhu) | 30.230595 111.677649 | [12] |
| AF306613-306621 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Wangjia Dayuan, Miao River (near Changhu) | 30.22455 111.754768 | [12] |
| AF306622-306630 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Songzi, Huayuanwu, Miao River (near Changhu) | 30.24713 111.742787 | [12] |
| DQ112267 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Jiangling, Xiaojiaju (Yangtze River) | 30.009 112.579 | [8] |
| DQ112268 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Jiangling, Xiaojiaju (Yangtze River) | 30.009 112.579 | [8] |
| EU001660 | Oncomelania hupensis hupensis | Hubei, Yudian River, Wuhan, Suizhou, Huangjiawan | 31.839056 113.681889 | [13] |
| EU333403 | Oncomelania hupensis hupensis | Wuhan, Jingmen,Liujialing (near Nanhu lake & Hanshui river) | 31.1 112.433333 | [10] |
| FJ997214 | Oncomelania hupensis hupensis | Hunan, Yueyang, Zhongzhouxiang (Xiajiang river) | 29.0861 113.0595 | [14] |
| GU367390 | Oncomelania hupensis hupensis | Hunan, Yue Yang, Linxiang, Baiyun | 29.483333 113.4 | [10] |
| GU367392 | Oncomelania hupensis hupensis | Hubei, Yichang, Yuan An Xian, Xiangjiapo (Juhe river) | 31.05 111.633333 | [10] |
| JF284689 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Shizikouzhen, Qunxing | 30.0127 111.9602 | unpub |
| JF284690 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Jiangling, Gejiatai, Zhujianao | 30.2261 112.4206 | unpub |
| JF284692 | Oncomelania hupensis hupensis | Hunan, Yue Yang, Zhongzhouxiang (Xiajiang river) | 29.0861 113.0595 | unpub |
| Mid-Yangtze Poyang Lake Basin (China)—PLB | ||||
| KR002674 | Oncomelania hupensis hupensis | Jiangxi, Jiangshan, Xianyanzhen, Houcang | 28.590853,118.370476 | |
| DQ112255-112263 | Oncomelania hupensis hupensis | Jiangxi, Jiujiang, Pengze (Yangtze river/Huanghu near Poyang Lake) | 28.983 116.15 | [9] |
| GU367393 | Oncomelania hupensis hupensis | Zhejiang, Quzhou, Kaihua, Chihuaizhen, Youchuan Cun (Huangshan) | 29.116667 118.2 | [10] |
| JF284693 | Oncomelania hupensis hupensis | Jiangxi, Jiujiang, Jingdezhen, Zhongshanpo (Daming Lake) | 28.9931 116.4887 | unpub |
| JF284694 | Oncomelania hupensis hupensis | Zhejiang, Quzhou, Jinhua, Liugoukou, Shafan Reservoir | 28.8523 119.4688 | unpub |
| Mid-Yangtze to Lower Yangtze Taihu Plain (China)—THP | ||||
| AF254490-254499 | Oncomelania hupensis hupensis | Anhui, Ning Guo County | 30.3730 118.9725 | [11] |
| AF254500-254509 | Oncomelania hupensis hupensis | Anhui, Chizhou, Guichi, Chikou | 30.6734 117.4549 | [11] |
| AF254510 | Oncomelania hupensis hupensis | Zhejiang, Guangde, Guzhushan, Houchong (near Taihu) | 31.0931 119.685 | [11] |
| AF254511 | Oncomelania hupensis hupensis | Zhejiang, Guangde, Guzhushan, Houchong (near Taihu) | 31.0931 119.685 | [11] |
| AF254512 | Oncomelania hupensis hupensis | Zhejiang, Guangde, Guzhushan, Houchong (near Taihu) | 31.0931 119.685 | [11] |
| AF254513 | Oncomelania hupensis hupensis | Zhejiang, Guangde, Guzhushan, Houchong (near Taihu) | 31.0931 119.685 | [11] |
| AF254514 | Oncomelania hupensis hupensis | Zhejiang, Guangde, Guzhushan, Houchong (near Taihu) | 31.0931 119.685 | [11] |
| AF254515 | Oncomelania hupensis hupensis | Zhejiang, Guangde, Guzhushan, Houchong (near Taihu) | 31.0931 119.685 | [11] |
| AF254516-254525 | Oncomelania hupensis hupensis | Anhui, Guangde County, Dongjia Datang, Xinhangzhen | 31.0490 119.550 | [11] |
| AF254526 | Oncomelania hupensis hupensis | Jiangsu, XiaShesizu, WangTaosizu, Dongtai, Yancheng, coastal | 32.8911 120.6460 | [11] |
| AF254527 | Oncomelania hupensis hupensis | Jiangsu, XiaShesizu, WangTaosizu, Dongtai, Yancheng, coastal | 32.8911 120.6460 | [11] |
| AF254528 | Oncomelania hupensis hupensis | Jiangsu, XiaShesizu, WangTaosizu, Dongtai, Yancheng, coastal | 32.8911 120.6460 | [11] |
| AF254529 | Oncomelania hupensis hupensis | Jiangsu, XiaShesizu, WangTaosizu, Dongtai, Yancheng, coastal | 32.8911 120.6460 | [11] |
| AF254530 | Oncomelania hupensis hupensis | Jiangsu, XiaShesizu, WangTaosizu, Dongtai, Yancheng, coastal | 32.8911 120.6460 | [11] |
| AF254531 | Oncomelania hupensis hupensis | Jiangsu, XiaShesizu, WangTaosizu, Dongtai, Yancheng, coastal | 32.8911 120.6460 | [11] |
| AF254532 | Oncomelania hupensis hupensis | Zhejiang, Huzhou, Guangde, Xianshanhu, Si'anzhen | 30.8955 119.6562 | [11] |
| AF254533 | Oncomelania hupensis hupensis | Zhejiang, Huzhou, Guangde, Xianshanhu, Si'anzhen | 30.8955 119.6562 | [11] |
| AF254534 | Oncomelania hupensis hupensis | Zhejiang, Huzhou, Guangde, Xianshanhu, Si'anzhen | 30.8955 119.6562 | [11] |
| AF254535 | Oncomelania hupensis hupensis | Zhejiang, Huzhou, Guangde, Xianshanhu, Si'anzhen | 30.8955 119.6562 | [11] |
| AF254536 | Oncomelania hupensis hupensis | Anhui, Tongling, Hujiamen, Guanghui Cun, LaoDao Island | 30.9542 117.7664 | [11] |
| AF254537 | Oncomelania hupensis hupensis | Anhui, Tongling, Hujiamen, Guanghui Cun, LaoDao Island | 30.9542 117.7664 | [11] |
| AF254538 | Oncomelania hupensis hupensis | Anhui, Tongling, Hujiamen, Guanghui Cun, LaoDao Island | 30.9542 117.7664 | [11] |
| AF254539 | Oncomelania hupensis hupensis | Anhui, Tongling, Hujiamen, Guanghui Cun, LaoDao Island | 30.9542 117.7664 | [11] |
| AF254540 | Oncomelania hupensis hupensis | Anhui, Tongling, Hujiamen, Guanghui Cun, LaoDao Island | 30.9542 117.7664 | [11] |
| AF254541 | Oncomelania hupensis hupensis | Anhui, Tongling, Hujiamen, Guanghui Cun, LaoDao Island | 30.9542 117.7664 | [11] |
| AF254542-254555 | Oncomelania hupensis hupensis | Anhui, Xuancheng, Sunbuzhen, Mawang Cun | 30.8780 118.9143 | [11] |
| JF284686 | Oncomelania hupensis hupensis | Anhui, Huzhou, Guangde, Yinjiwan, Jingliu Cun (Taihu lake) | 31.0675 119.4432 | unpub |
| JF284687 | Oncomelania hupensis hupensis | Anhui, Guangde, Shanbeixiang, Jishan Cun, Nianzhiwu | 31.0675 119.4432 | unpub |
| JF284688 | Oncomelania hupensis hupensis | Jiangsu, Yangzhou, Wantou (Yangtze river) | 32.2651 119.5707 | unpub |
| Sichuan: Anning River Valley (China)—SAV | ||||
| AF213339 | Oncomelania hupensis robertsoni | Liangshan, Shizuizi | 27.84284 102.39277 | [15] |
| DQ112250 | Oncomelania hupensis robertsoni | Liangshan, Miyi, Hongshiyan, Guanyinxiang, Shujingwan | 26.963611 102.132778 | [9] |
| DQ112251 | Oncomelania hupensis robertsoni | Liangshan, Miyi, Hongshiyan, Guanyinxiang, Shujingwan | 26.963611 102.132778 | [9] |
| DQ212814-212820 | Oncomelania hupensis robertsoni | Liangshan, Xixiangxiang, Gucheng Cun | 27.93525 102.20540 | [8] |
| DQ212821 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] |
| DQ212822 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] |
| DQ212823 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] |
| DQ212824 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] |
| DQ212825 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] |
| DQ212826 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] |
| DQ212827 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] |
| DQ212828 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] |
| DQ212829 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang | 27.7995 102.3087 | [8] |
| DQ212830 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang1 | 27.7995 102.3087 | [8] |
| DQ212831 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang1 | 27.7995 102.3087 | [8] |
| DQ212832 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang1 | 27.7995 102.3087 | [8] |
| DQ212833 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang2 | 27.7973 102.3157 | [8] |
| DQ212834 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang2 | 27.7973 102.3157 | [8] |
| DQ212835 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang2 | 27.7973 102.3157 | [8] |
| DQ212836 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang2 | 27.7973 102.3157 | [8] |
| DQ212837 | Oncomelania hupensis robertsoni | Liangshan G 5 Jing Kun Gao Su Chu Kou, Dafenba | 27.7468 102.1903 | [8] |
| DQ212838 | Oncomelania hupensis robertsoni | Liangshan G 5 Jing Kun Gao Su Chu Kou, Dafenba | 27.7468 102.1903 | [8] |
| DQ212839 | Oncomelania hupensis robertsoni | Liangshan G 5 Jing Kun Gao Su Chu Kou, Dafenba | 27.7468 102.1903 | [8] |
| DQ212840 | Oncomelania hupensis robertsoni | Liangshan G 5 Jing Kun Gao Su Chu Kou, Dafenba | 27.7468 102.1903 | [8] |
| DQ212841-212851 | Oncomelania hupensis robertsoni | Liangshan, Miyi, G5 Jingkun expressway, Shujingwan | 26.9637 102.1328 | [8] |
| EU079378 | Oncomelania hupensis robertsoni | Liangshan, Mazengyiwuxiang, Weija Diaolou | 27.817833 102.369472 | [13] |
| JF284691 | Oncomelania hupensis robertsoni | Liangshan, Erwu, Yiwanshui | 27.8773 102.3703 | unpub |
| Sichuan Plain (China)—SCB | ||||
| KR002675 | Oncomelania hupensis robertsoni | Chengdu Xinjin | 30.405452,103.829727 | |
| AF531547 | Oncomelania hupensis robertsoni | Mianzhu, Tiezhuangyan (Mawei River) | 31.302792 104.215357 | [16] |
| DQ212797 | Oncomelania hupensis robertsoni | Meishan, Danling, Xindianzi, Qinglongzui | 29.99163 103.41580 | [8] |
| DQ212798 | Oncomelania hupensis robertsoni | Meishan, Danling, Xindianzi, Qinglongzui | 29.99163 103.41580 | [8] |
| DQ212799-212808 | Oncomelania hupensis robertsoni | Meishan Dongpo Pan'aoxiang Wangshigou | 30.13993 103.61167 | [8] |
| DQ212809 | Oncomelania hupensis robertsoni | Meishan, Dongpo, Baiyang Cun | 30.0373 103.9002 | [8] |
| DQ212810 | Oncomelania hupensis robertsoni | Meishan, Dongpo, Baiyang Cun | 30.0373 103.9002 | [8] |
| DQ212811 | Oncomelania hupensis robertsoni | Meishan, Dongpo, Baiyang Cun | 30.0373 103.9002 | [8] |
| DQ212812 | Oncomelania hupensis robertsoni | Meishan, Dongpo, Baiyang Cun | 30.0373 103.9002 | [8] |
| DQ212813 | Oncomelania hupensis robertsoni | Meishan, Dongpo, Baiyang Cun | 30.0373 103.9002 | [8] |
| JF284697 | Oncomelania hupensis robertsoni | Ya'an, Qionglai, Daxingzhen, Shuikou Cun, Xucitang | 30.2701 103.4562 | unpub |
| Taiwan (China)—TAI | ||||
| DQ112271-112280 | Oncomelania hupensis chiui | Taipei, Shimen, Alilao Cun (Coastal) | 25.278607 121.61164 | unpub |
| DQ112281 | Oncomelania hupensis formosana | Taiwan (W) Changhua, Puyan Township | 23.996416 120.475913 | [8] |
| DQ112282 | Oncomelania hupensis formosana | Taiwan (W) Changhua, Puyan Township | 23.996416 120.475913 | [8] |
| DQ112283 | Oncomelania hupensis formosana | Taiwan (W) Changhua, Puyan Township | 23.996416 120.475913 | [8] |
| EF394891 | Oncomelania hupensis chiui | Taipei, Shimen, Alilao Cun (Coastal) | 25.279121 121.611056 | [17] |
| Yunnan: Erhai Basin—YEB | ||||
| AF253074 | Oncomelania hupensis robertsoni | Dali, Tongsipan | 25.4517 100.2011 | unpub |
| AF253075 | Oncomelania hupensis robertsoni | Dali, Tongsipan | 25.4517 100.2011 | unpub |
| DQ112252 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ112253 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ112254 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ212852 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ212853 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ212854 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ212855 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ212856 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| DQ212857 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] |
| EU688958 | Oncomelania hupensis robertsoni | Dali, Nanjianzhen | 25.043391 100.53031 | unpub |
| EU688959 | Oncomelania hupensis robertsoni | Lijiang, Yongsheng | 26.664488 100.758963 | unpub |
| EU780225 | Oncomelania hupensis robertsoni | Dali, Chao Yang Cun | 25.82887 100.11777 | unpub |
| GU367389 | Oncomelania hupensis robertsoni | Lijiang, Yongsheng | 26.683333 100.75 | unpub |
| Japan—JAP | ||||
| KR002673 | Oncomelania hupensis nosophora | Honshu, Chiba Kisarazu | 35.350371 139.988407 | |
| AB611787 | Oncomelania hupensis nosophora | Honshu, Yamanashi, Nirasaki | 35.715 138.434 | [18] |
| AB611791 | Oncomelania minima | Honshu, Ishikawa, Wajima | 37.376 136.894 | [18] |
| AB611795 | Oncomelania minima | Sado Island, Niigata | 38.014 138.368 | [18] |
| DQ112284 | Oncomelania hupensis nosophora | Honshu Kanagawa Kiyokawa | 35.3976 139.9533 | [8] |
| DQ112285 | Oncomelania hupensis nosophora | Honshu Kanagawa Kiyokawa | 35.3976 139.9533 | [8] |
| DQ112286 | Oncomelania hupensis nosophora | Honshu Kanagawa Kiyokawa | 35.3976 139.9533 | [8] |
| Philippines—PHL | ||||
| JF284698 | Oncomelania hupensis quadrasi * | Mindoro, Mansalay | 12.547 121.442 | [19] |
| DQ112289 | Oncomelania hupensis quadrasi | Leyte, Palo | 11.16 124.98 | unpub |
| DQ112287 | Oncomelania hupensis quadrasi | Leyte, Palo | 11.16 124.98 | unpub |
| DQ112288 | Oncomelania hupensis quadrasi | Leyte, Palo | 11.16 124.98 | unpub |
Data are: GenBank accession numbers (GACs), collection localities (with coordinates and region code as used in Fig 1, e.g., FCP) and (Ref) source references (citations in italics refer to locality information only and not to the publication of the GAC; unpub, indicates unpublished data with locations drawn from isolate codes, specimen voucher numbers or details in the GenBank). Taxonomic nomenclature follows[9]. Data collected as part of the present study are marked in bold.
*Incorrectly listed as Chinese Oncomelania hupensis hupensis on GenBank
Table 2. Malacological data used in the phylogeographical reconstructions–rrnL locus (16S).
| GAC | Taxon | Locality | Coordinates | Ref | ||
|---|---|---|---|---|---|---|
| Fujian coastal plain (China)—FCP | ||||||
| DQ212860 | Oncomelania hupensis tangi | Fuzhou, Fuqing, Donggang, Donghanzhen | 25.430833, 119.605278 | [8] | ||
| JF284695 | Oncomelania hupensis tangi * | Fuzhou, Fuqing, Donggang, Donghanzhen, Nansha | 25.4049 119.6471 | unpub | ||
| Guangxi coastal plain (China)—GCP | ||||||
| JF284696 | Oncomelania hupensis | Nanning, Jing Xi Xian, Baise, Bameng Reservoir | 23.3655 106.2997 | unpub | ||
| Hunan / Hubei Dongting Lake Basin (China)—DLB | ||||||
| EU001660 | Oncomelania hupensis hupensis | Hubei, Yudian River, Wuhan, Suizhou, Huangjiawan | 30.189056 112.181889 | [13] | ||
| FJ997214 | Oncomelania hupensis hupensis | Hunan, Yueyang, Zhongzhouxiang (Xiajiang river) | 29.0861 113.0595 | [14] | ||
| JF284689 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Shizikouzhen, Qunxing | 30.0127 111.9602 | unpub | ||
| JF284690 | Oncomelania hupensis hupensis | Hubei, Jingzhou, Jiangling, Gejiatai, Zhujianao | 30.2261 112.4206 | unpub | ||
| JF284692 | Oncomelania hupensis hupensis | Hunan, Yue Yang, Zhongzhouxiang (Xiajiang river) | 29.0861 113.0595 | unpub | ||
| Mid-Yangtze Poyang Lake Basin (China)—PLB | ||||||
| JF284687 | Oncomelania hupensis hupensis | Anhui, Guangde, Shanbeixiang, Jishan Cun, Nianzhiwu | 31.0675 119.4432 | unpub | ||
| JF284693 | Oncomelania hupensis hupensis | Jiangxi, Jiujiang, Jingdezhen, Zhongshanpo (Daming Lake) | 28.9931 116.4887 | unpub | ||
| JF284694 | Oncomelania hupensis hupensis | Zhejiang, Quzhou, Jinhua, Liugoukou, Shafan Reservoir | 28.8523 119.4688 | unpub | ||
| Mid-Yangtze to Lower Yangtze Taihu Plain (China)—THP | ||||||
| DQ21259 | Oncomelania hupensis hupensis | Wuhu, Xuancheng, Ningguo, Jiangjiashan | 30.641921 118.841993 | [8] | ||
| JF284686 | Oncomelania hupensis hupensis | Anhui, Huzhou, Guangde, Yinjiwan, Jingliu Cun (Taihu lake) | 31.0675 119.4432 | unpub | ||
| JF284688 | Oncomelania hupensis hupensis | Jiangsu, Yangzhou, Wantou (Yangtze river) | 32.2651 119.5707 | unpub | ||
| Sichuan: Anning River Valley (China)—SAV | ||||||
| AF212893 | Oncomelania hupensis robertsoni | Liangshan, Shizuizi | 27.84284 102.39277 | [15] | ||
| DQ212872 | Oncomelania hupensis robertsoni | Liangshan, Xixiangxiang, Gucheng Cun | 27.93525 102.20540 | [8] | ||
| DQ212873 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang | 27.7973 102.3157 | [8] | ||
| DQ212874 | Oncomelania hupensis robertsoni | Liangshan, Xixiangxiang, Gucheng Cun | 27.93525 102.20540 | [8] | ||
| DQ212875 | Oncomelania hupensis robertsoni | Liangshan, Xixiangxiang, Gucheng Cun | 27.93525 102.20540 | [8] | ||
| DQ212876 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] | ||
| DQ212877 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] | ||
| DQ212878 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] | ||
| DQ212879 | Oncomelania hupensis robertsoni | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | [8] | ||
| DQ212880 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] | ||
| DQ212881 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] | ||
| DQ212882 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] | ||
| DQ212883 | Oncomelania hupensis robertsoni | Liangshan off left side of G5 Jingku Gao Su, Jingjiuxiang | 27.8000 102.204 | [8] | ||
| DQ212884 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang1 | 27.7995 102.3087 | [8] | ||
| DQ212885 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang1 | 27.7995 102.3087 | [8] | ||
| DQ212886 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang1 | 27.7995 102.3087 | [8] | ||
| DQ212887 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang2 | 27.7973 102.3157 | [8] | ||
| DQ212888 | Oncomelania hupensis robertsoni | Liangshan Qionghai Lake, Hainanxiang2 | 27.7973 102.3157 | [8] | ||
| DQ212889 | Oncomelania hupensis robertsoni | Liangshan G 5 Jing Kun Gao Su Chu Kou, Dafenba | 27.7468 102.1903 | [8] | ||
| DQ212890 | Oncomelania hupensis robertsoni | Liangshan G 5 Jing Kun Gao Su Chu Kou, Dafenba | 27.7468 102.1903 | [8] | ||
| DQ212891-212898 | Oncomelania hupensis robertsoni | Liangshan, Miyi, G5 Jingkun expressway, Shujingwan | 26.9637 102.1328 | [8] | ||
| EU079378 | Oncomelania hupensis robertsoni | Liangshan, Mazengyiwuxiang, Weija Diaolou | 27.817833 102.369472 | [13] | ||
| JF284691 | Oncomelania hupensis robertsoni | Liangshan, Erwu, Yiwanshui | 27.8773 102.3703 | unpub | ||
| Sichuan Plain (China)—SCB | ||||||
| KR002677 | Oncomelania hupensis robertsoni | Chengdu Xinjin | 30.405452 103.829727 | |||
| AF531545 | Oncomelania hupensis robertsoni | Mianzhu, Tiezhuangyan (Mawei River) | 31.302792 104.215357 | [16] | ||
| DQ212863-212871 | Oncomelania hupensis robertsoni | Meishan Dongpo Pan'aoxiang Wangshigou | 30.1399 103.6117 | [8] | ||
| JF284697 | Oncomelania hupensis robertsoni | Ya'an, Qionglai, Daxingzhen, Shuikou Cun, Xucitang | 30.2701 103.4562 | unpub | ||
| Taiwan (China)—TAI | ||||||
| DQ212861 | Oncomelania hupensis formosana | Taiwan (W) Changhua, Puyan Township | 23.996416 120.475913 | [8] | ||
| Yunnan: Erhai Basin—YEB | ||||||
| DQ212899 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] | ||
| DQ212900 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] | ||
| DQ212901 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] | ||
| DQ212902 | Oncomelania hupensis robertsoni | Dali, Yongjianzhen, Tongsipan | 25.4510 100.2007 | [8] | ||
| Japan—JAP | ||||||
| KR002676 | Oncomelania hupensis nosophora | Honshu, Chiba Kisarazu | 35.350371 139.988407 | |||
| AB611786 | Oncomelania hupensis nosophora | Honshu, Yamanashi, Nirasaki | 35.715 138.434 | [18] | ||
| AB611790 | Oncomelania minima | Honshu, Ishikawa, Wajima | 37.376 136.894 | [18] | ||
| AB611794 | Oncomelania minima | Sado Island, Niigata | 38.014 138.368 | [18] | ||
| DQ212858 | Oncomelania minima | Honshu, Tokyo Chiyoda | 35.6862 139.7534 | [8] | ||
| Philippines—PHL | ||||||
| JF284698 | Oncomelania hupensis quadrasi * | Mindoro, Mansalay | 12.547 121.442 | unpub | ||
| DQ212862 | Oncomelania hupensis quadrasi | Luzon, Tarlac, Victoria, Calibungan | 15.5930 120.7388 | [8] | ||
Details as Table 1.
*Incorrectly listed as Chinese Oncomelania hupensis hupensis on GenBank
Table 3. Parasite data used in the phylogeographical reconstructions.
| GAC | Sampling method | Locality | Coordinates | Ref | ||
|---|---|---|---|---|---|---|
| Hunan / Hubei Dongting Lake Basin(China)—DLB | ||||||
| HM120842 | FCC | Hubei, Wuhan, Caidian, Beiban (Houguan Hou) | 30.569 114.017 | [20] | ||
| HM120845 | FCC | Hunan, Yue Yang, Junshan, Tangang Cun (Yangtze River) | 29.617 113.067 | [20] | ||
| Mid-Yangtze Poyang Lake Basin (China)—PLB | ||||||
| HM120844 | FCC | Jiangxi, Jiujiang, Jingdezhen, Zhongshanpo (Daming Lake) | 28.9931 116.4887 | [20] | ||
| Mid-Yangtze to Lower Yangtze Taihu Plain (China)—THP | ||||||
| HM120843 | FCC | Zhejiang, Guangde, Guzhushan, Houchong (near Taihu) | 31.0931 119.6850 | [20] | ||
| NC002544 | ADL | Anhui, Tongling, Hujiamen, Guanghui Cun, LaoDao Island | 30.9542 117.7664 | [21] | ||
| KF279405-6 | ADL | Anhui, Chizhou, Guichi, Chikou | 30.6734 117.4549 | unpub | ||
| HM120841 | FCC | Anhui, Guangde, Shanbeixiang, Jishan Cun, Nianzhiwu | 31.0675 119.4432 | [20] | ||
| Sichuan: Anning River Valley (China)—SAV | ||||||
| KF279407 | ADL | Liangshan (near Qionghai sea) Yanjia Buzi (Heyan Road) | 27.87505 102.30867 | unpub | ||
| Sichuan Plain (China)—SCB | ||||||
| HM120846 | FCC | Meishan, Dongpo, Baiyang Cun | 30.0373 103.9002 | [20] | ||
| Taiwan—TAI | ||||||
| KF279410 | Taiwan (W) Changhua, Puyan Township | 23.996416 120.475913 | unpub | |||
| Yunnan: Erhai Basin—YEB | ||||||
| HM120847-8; KF279408-9 | FCC | Yunnan, Xizhou Chaoyang Cun; Yunnan, Xizhou, Sili Cun | 25.82887 100.117770 | [20]; unpub | ||
| Japan—JAP | ||||||
| JQ781215 | ADL | Honshu, Yamanashi, Nirasaki | 35.715 138.434 | unpub | ||
| Philippines—PHL | ||||||
| JQ781211-12 | ADL | Mindoro, Mansalay | 12.547 121.442 | unpub | ||
| Outgroup: Schistosoma mekongi | ||||||
| NC002529 | ADL | Lao PDR, Khong District, Ban Hat-Xai-Kuhn (Mekong River) | 14.12056 105.865860 | [21] | ||
Loci sampled are cox1 and cox3 (extracted from longer contigs, including complete mitochondrial genomes). Data are: GenBank accession numbers (GACs), collection localities (with coordinates) and source references; unpub, indicates unpublished data with locations drawn from isolate codes, specimen voucher numbers or details in the GenBank. ADL indicates adult worms from laboratory lines based on field-collected infected snails, FCC stands for cercariae from snails collected in the field and used directly to infect laboratory hosts.
After submission of this manuscript, new data for Philippine and Japanese Schistosoma japonicum were added to the GenBank by unpublished authors of another laboratory. These data are included in Table 3, but were not included in the hypothesis testing; nevertheless, a phylogeny was estimated using this extended data set in order to assess the impact of the new data on the conclusions of this study (if any).
Initial handling of data and selection of partitioning scheme and substitution models
Sequence data were extracted from GenBank using Biopython 1.61 Bio.SeqIO[44]. The sequences were aligned using MUSCLE 3.8.31[45], with default settings. To reduce computing time in subsequent analyses, the alignments were inspected in SeaView 4.4.2[46] and the first 240 and last 60 bps of the complete cox1 gene for the S. japonicum (worm) alignment were excluded (the ingroup showed no variation in these regions). Taxa with identical sequences at a locus (gene) were then excluded, leaving one representative of each haplotype: for the worms identical sequences only occurred within villages or townships, but for the snails identical sequences were found up to county level. In data sets where sequences for two loci were concatenated, data were only excluded if sequences were identical between two taxa at both loci. The final data set for the worms comprised a concatenated cox1+cox3 sequence, and had 16 taxa (19 in the extended data set) and 1994 characters (after removal of 2 identical sequences). As cox1 and rrnL data were not both available for all snail taxa, there were two data sets for the snails. The cox1 data set comprised 146 taxa and 599 characters (after removal of 129 identical sequences) and a second data set was made using pyfasta 0.5.2 to select all cox1 sequences for which there was a corresponding rrnL sequence; these sequences were then concatenated to produce a cox1+rrnL “both loci” alignment of 51 taxa and 1110 characters. The reading frame of the protein coding loci was determined using ExPASy Translate[47].
In addition to the alignments for individual sequences, population level data sets were produced for the snails because these were expected to be easier to visualise and detect dispersal tracts. To achieve this, the geographical range of the samples was divided into 10 biogeographical regions (see Fig 1) such that within each region there was no barrier to dispersal of Oncomelania; thus there would be only isolation by distance and no major ecological (e.g., lack of suitable habitat) or physical (e.g., mountain ranges or ridges) barriers; however it should be noted that the Tai Hu Plain (THP) region is likely to be interrupted by industrialised belts where there are no Oncomelania habitats. In contrast, each region is separated by mountains or similar areas of highland or sea. Consensus sequences were produced for the individuals within each region and a population sequence alignment estimated. The population data set included 13 ingroup taxa (JAP has two island populations, and the Philippines (PHL) was also included).
Phylogenetic analysis was performed using two fundamentally different approaches; the non-parametric Maximum Parsimony (MP) approach and the parametric Maximum Likelihood approach. Two contrasting methods were used so that resilience of the hypothesis to changes in method and associated assumptions could be used to reveal weakly supported clades. The rationale was that any clade that was represented in phylogenies found by both methods (and well supported) would be a reliable reconstruction of phylogenetic history for these taxa. In addition, the strategy helps reveal artefacts associated with a specific class of methods.
Phylogenetic analysis by Maximum Likelihood (ML) with RAxML
RAxML 7.4.8[48] was chosen to implement the ML analysis because, among currently available programs, RAxML has shown best performance in terms of inference times and final likelihood values, and has a rapid boot-strapping algorithm which allows clade support to be estimated in reasonable time frames, even when estimating null distributions. A series of test runs were used to determine optimum values or states for the settings of the RAxML analyses (details published elsewhere[30]). The apparently optimum partitioning strategy and evolutionary model for each partition was determined using PartitionFinder 1.0.1[49], under a BIC criterion and models restricted to those implementable in RaxML. The chosen models for the snails were: cox1, GTR+G codons CP123; cox1+rrnL, GTR+G codons CP123 and rrnL separately partitioned; populations, GTR+G codons CP111 cox1 and rrnL partitioned separately. For the worms the models were, for cox1+cox3, GTR+G codons CP112 with cox1 and cox3 codons in the same partition (i.e., there were two only partitions). RAxML was run with 100000 bootstrap replicates, using every fifth tree found by bootstrapping as a starting tree for a series of 20,000 full ML searches. The CAT approximation in RAxML was not used. Three main runs were performed with different random number seeds. After checking that the independent runs led to trees of the same topology and very similar levels of support (±1%), the bootstrap trees for all three runs were pooled and a 100% (strict) majority rule consensus tree reported.
For the hypothesis testing, a data set was constructed that included all snail taxa for which there was a worm sample taken at the same locality. These data included ten taxa and 1994 characters. The model and partition scheme for the worms was the same as for the full worm data set, and for the snails a two partition model was again chosen: GTR+G (rrnL, cox1codon1, cox1codon2) and cox1codon3.
Phylogenetic analysis by Maximum Parsimony (MP) with POY
POY 5.1.1[50] (parallelised build) was used to implement a Maximum Parsimony approach. The use of MP afforded an analysis free of the assumptions underlying ML methods, and the use of POY with its dynamic homology approach (where characters (transformation series) are inferred during the process of phylogenetic reconstruction) frees the analysis of any effects particular to the alignment inferred by MUSCLE[51]. A sensitivity analysis was used to choose the weighting (gap cost etc) and partition schemes for each data set, protocol published elsewhere[52].
Hypothesis testing
In order to test whether the evolution of the DNA sequences was consistent with a particular hypothesis, such as coevolution of Oncomelania hupensis and Schistosoma japonicum, the Incongruence Length Difference test[53] (ILD) was used as implemented in PAUP* 4.0b10[54], with 5000 replicates. The test employed a sub-set of the data which included only polymorphic sites (for reasons published elsewhere[55]). The test, which randomly exchanges segments between the snail and worm data partitions, should give ML outcomes which are not significantly different from those of the original data if the snails and worms evolved to a common history. The ILD has been shown to be rather conservative when used as a test of topological congruence if phylogenies with trees of similar topologies are compared, with the opposite effect observed if the trees compared differ markedly in structure (e.g., internal branch length differences)[56,57]. Noise in the phylogenetic signal can also lead to type-I errors in the ILD test[58]. Consequently, the hypotheses were also tested using Monte Carlo simulation in the manner of the SOWH test[59]. In the case of the test for coevolution, the test statistic is the likelihood ratio of the phylogeny inferred for the worm data (unconstrained) and the same phylogeny inferred with the evolutionary history constrained to that of the snails (represented by the ML tree estimated for the snail data). A null distribution of the test statistic was calculated by simulating many data sets using Seq-Gen 1.3.3–1[60] and the ML parameters of the substitution model inferred for the worm data, but constrained under the null hypothesis (the ML tree for the original worm data constrained by the snail ML tree). For each simulacrum the ML tree was estimated both unconstrained and constrained by the snail ML tree, and a likelihood ratio computed. The null distribution then being a distribution for the amount of discord expected to occur when the worm phylogeny had evolved according to the same history as the snails. If the likelihood ratio for the original data falls above the 95th percentile of the null distribution, the hypothesis that both worms and snails evolved to the same (i.e., the worms') phylogenetic history can be rejected at P<0.05. The replicates were performed using RAxML (with settings as for the original worm data, but bootstrapping set to terminate according to a convergence criterion based on the extended majority rule consensus trees), and they continued until the null distribution appeared to have stabilised, as judged by a plateau of t-values with increasing replicate number.
Visualisation of phylogenies and phylogeographies
In cases where the topologies resulting from phylogenetic analysis by ML and MP did not agree for a particular data set, a strict consensus tree was generated from the two trees so that discordant clades were represented by polytomies. Consequently, the resolved clades in the final trees for each data set were those supported by both methods. In order to visualise the phylogeographies of both snails and worms, the phylogenies were mapped in Marble Virtual Globe 1.8.3 using the Phylo2GE R script. In addition, phylogenies were compared (topologically) using the compare2Trees algorithm[61], which scores each pair of branches, between the trees, according to the common taxon partitions they define, with the branches then paired so as to maximise the overall score; this process yields a score (S) value for a pair of trees.
Results
Phylogenetic reconstructions
To enable RAxML and reduce computing time 129 identical and/or ambiguous cox1 sequences were excluded from the original snail data set (i.e., the sequence alignment for Oncomelania) to leave 146 taxa. Only one of the 129 exclusions involved identical sequences at the inter-regional scale. The haplotype diversity by region was roughly inversely proportional to the sampling intensity (i.e., number of sequences per region), except for YEB and PHL, and to a lesser degree SCB, which had small sample sizes and low haplotype diversities (Table 4).
Table 4. Numbers of individuals sequenced (N) and haplotype diversity (H) by region for the Cox1 and rrnL data sets (snails) and the Cox1+Cox3 data set (worms).
| Cox1 | rrnL | Cox1+Cox3 | |||
|---|---|---|---|---|---|
| Region | N | H | N | H | N |
| FCP | 2 | 1.00 | 2 | 1.00 | 0 |
| GCP | 5 | 0.80 | 1 | 1.00 | 0 |
| DLB | 74 | 0.55 | 5 | 0.80 | 2 |
| PLB | 12 | 0.92 | 3 | 1.00 | 1 |
| THP | 69 | 0.45 | 3 | 1.00 | 5 |
| SAV | 43 | 0.47 | 32 | 0.84 | 1 |
| SCB | 20 | 0.50 | 12 | 0.75 | 1 |
| TAI | 14 | 0.71 | 1 | 1.00 | 1 |
| YEB | 15 | 0.47 | 4 | 0.50 | 4 |
| JAP | 7 | 0.86 | 5 | 1.00 | 0 |
| PHL | 4 | 0.50 | 2 | 1.00 | 0 |
The haplotype diversity for the worms was 1.00 for all regions. Region codes are described in Fig 1.
Replicate phylogenetic analyses run for the snail data, with different random seeds and only the cox1 data, failed to result in a common tree (S<0.88), and there was poor agreement between the results of the ML and MP searches (S<0.55); the phylogeny also contained many unresolved clades. Consequently, the cox1 data were considered to lack phylogenetic signal and were not considered further in this study. In contrast, the phylogenies estimated for both loci (cox1 and rrnL concatenated) showed good agreement among replicate runs (S>0.92) and between ML and MP (S>0.75).
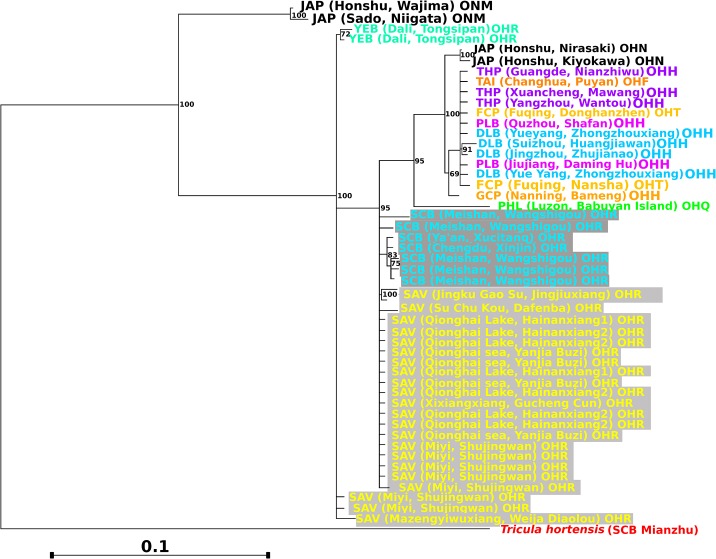
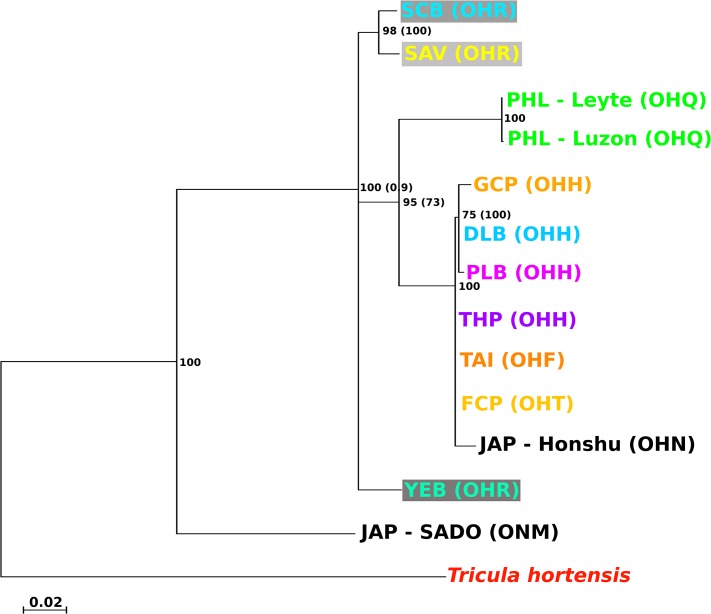
The strict (100%) consensus tree between the ML and MP searches is given in Fig 2. Considering O. hupensis, the only biogeographical regions characterised as monophyletic clades are JAP and YEB. Nevertheless, the Sichuan populations (SCB and SAV), which lie in an isolated mountainous area, form an unresolved near-monophyletic clade, except for three SAV individuals which form a further unresolved clade at the base of the O. hupensis clade (this could be a result of long branch attraction due to saturation or a lack of apomorphies in younger clades[62], with slippage of long branches leading to SAV taxa, towards the root of the tree). The YEB populations, also mountainous but separated from the Sichuan populations by the Hengduan Range, formed a distinct basal clade at the same level as the three extraneous SAV samples. Thus, the basal clades of the phylogeny are occupied solely by O. h. robertsoni (and O. minima). The remaining major clade, which appears derived from O. h. robertsoni contains all the other O. hupensis samples, including the non-Chinese taxa O. h. quadrasi (Philippines), which is basal to the clade, and O. h. nosophora (Japan)–suggesting that these taxa did not diverge in situ. Although O. h. nosophora is monophyletic and O. h. robertsoni is near monophyletic (it's apparent polyphyly might be explained by long branch attraction), O. h. hupensis is polyphyletic and includes O. h. tangi and O. h. formosana (of FCP and Taiwan, respectively); this suggests that the latter two may be populations of O. h. hupensis. The GCP taxon lies at the base of the Chinese O. h. hupensis clade and this gives some support to the case for O. h. guangxiensis. The populations of the lake and marshland region (DLB, FCP and THP) form an unresolved clade, suggesting that there are few barriers to migration (gene-flow) between them. In view of this, the population phylogeny (where data for individuals is pooled to provide consensus sequences for each region) provides a representative summary of the phylogeny (Fig 3). The population phylogeny agrees with that for both loci, based on individuals, except that it shows GCP as forming a sub-clade, of the Lake and Marshland, Taiwan and Japan clade, with PLB and DLB.
Fig 2. Phylogram representing the strict (100%) consensus tree between Maximum Parsimony (MP) and Maximum Likelihood (ML) phylogenies.
As estimated for the Oncomelania populations sampled at both loci, cox1 and rrnL, with clade support values averaged from 100000 bootstrap replicates (ML) and 5000 jackknife iterations (MP). The colours assigned to the taxon names correspond to those of the biogeographical areas mapped in Fig 1. The light and dark grey shading denotes the western mountain clades. The outgroup is in red.
Fig 3. Population phylogeny for Oncomelania at both loci cox1 and rrnL.
Conditions of the analyses and colour scheme follow Fig 2, except that the DNA sequence data for all individuals were pooled into a consensus sequence for each biogeographical region before the sequences for the two loci were concatenated.
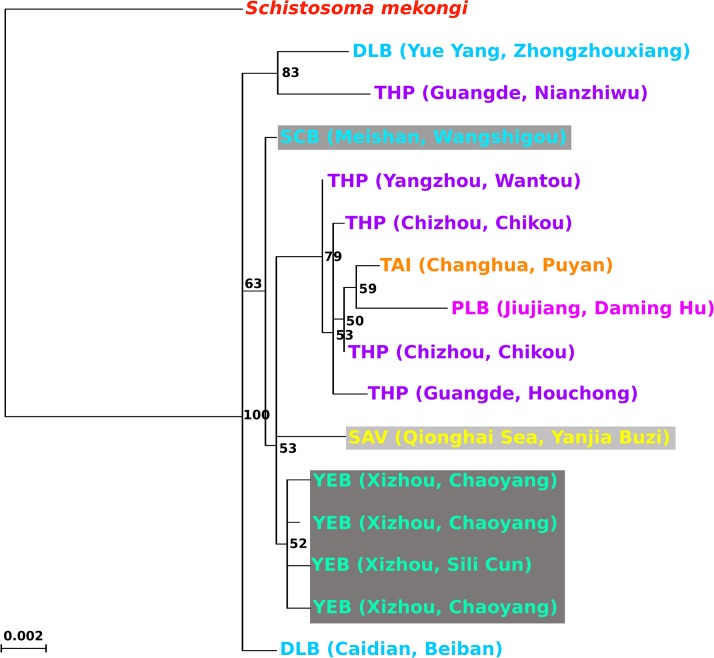
The phylogeny estimated for the worms (Fig 4) differed from that of the snails in certain key features. For example, some DLB and THP populations are basal to the western mountain clades (YEB, SCB and SAV). As in the snail phylogeny, the Taiwan population falls within a clade comprised solely of Lake and Marshland mainland Chinese taxa; however, this clade excludes DLB and so contains only THP, PLB and TAI, also the Taiwan sample clusters with PLB forming a clade derived from the Chikou THP samples.
Fig 4. Phylogeny estimated for Schistosoma japonicum.
Conditions of the analyses and colour scheme follow Fig 2.
Hypothesis testing
The snail and corresponding worm phylogeny (i.e., that estimated in exclusion of taxa not held in common; Fig 5) showed a low level of correspondence (S = 0.46). Consequently, it was necessary to test the null hypothesis that the snails and worms had evolved to a common evolutionary history, i.e., that of the snails. Initially the ILD test was used to detect significant conflict in the phylogenetic signals of the snail and worm data. The test was significant (P = 0.00004) suggesting that the two data partitions are the result of different evolutionary processes. To test further the null hypothesis, a SOWH test was performed; this resulted in a likelihood ratio for the observed data (unconstrained / constrained by the null hypothesis tree) of 75.52 and a 95th percentile for the null distribution of 4.26. Consequently, the null hypothesis can be rejected in the light of these data (P<0.0001). In view of these findings it appears highly unlikely that the evolutionary radiation of Schistosoma japonicum across China was shaped or driven by that of the snail intermediate hosts.
Fig 5. A comparison of the snail (left) and worm (right) population phylogenies.
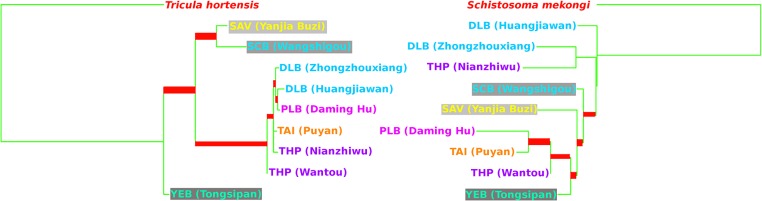
Clades which do not occur on both trees are marked in red (based on the output of compare2Trees). Conditions of the phylogenetic analyses and colour scheme follow Fig 2.
Phylogeography
Geospatially projected phylogenies (Fig 6 and Files A and B in S1 Dataset) assist in phylogeographical interpretation and in the present study they reveal clear differences between the radiations of the snails and the worms. The map for the snails (Fig 6A) suggests initial colonisation of the valleys of the western mountains (SAV, SCB, YEB) by a proto-Oncomelania hupensis robertsoni; thereafter these lineages, established in their respective valleys and basins, appear to have stabilised after initial divergence (no further cladogenesis), and remained so throughout most of the history of O. hupensis. The mountain clade appears to have given rise to the Lake and Marshland and East-coast clades (THP, FCP) of Chinese O. h. hupensis, with radiations back into Japan (as O. h. nosophora) and to Taiwan and the Philippines more recently in the history of this species. A second, slightly more recent, radiation from DLB, west to GCP and eastwards to PLB, then occurs. In contrast the worms show a history where the Lake and Marshland (eastern) populations appear to involve cladogenic events that occur throughout the history of Chinese S. japonicum, whereas the western mountain taxa are stable following initial establishment in their respective valleys. Unlike the snails, the worms show a most recent colonisation event of Taiwan that is associated with the PLB region rather than THP/FCP.
Fig 6. Projection of the phylogeny for the Oncomelania (A) and Schistosoma japonicum (B) populations onto a topographic map.
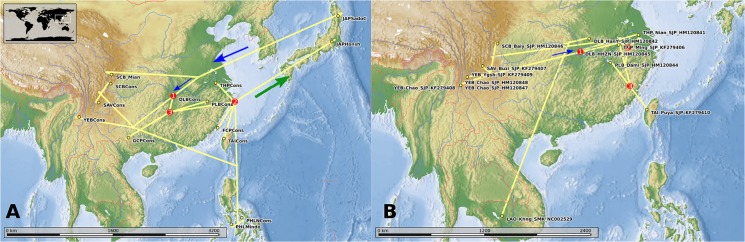
Plotted using Marble Virtual Globe (open source). The nodes in red indicate key divergence events and are numbered chronologically (see Discussion for details). The blue arrows show the polarity of initial radiation according to the East to West hypothesis and the green arrow a later back colonisation. For an interactive projection please use the kml files for these maps (S1 Dataset) provided in the Supporting Information.
After submission of this manuscript, new data for Philippine and Japanese Schistosoma japonicum were added to the GenBank. In order to determine the position of these additional populations in the phylogeography and their congruence with the snail phylogenies, an extended data set was subjected to phylogenetic analysis. File C in S1 Dataset and S1 Fig give the resulting kml and an image showing the extended phylogeny projected onto a globe, respectively.
Discussion
Taxonomy and phylogenetics
Of the four Chinese Oncomelania hupensis sub-species described by Davis[9], only two are supported in the present study. Oncomelania h. robertsoni is not polyphyletic and all individuals sampled of this sub-species are basal in the phylogeny; therefore, this taxon is supported. In contrast, O. h. hupensis is polyphyletic and includes O. h. tangi and O. h. formosana; this suggests that O. h. tangi and O. h. formosana are not valid sub-species, but are O. h. hupensis. Indeed O. hupensis populations can demonstrate considerable differences in morphology (mostly of the shell) and yet cluster together genetically[11]; therefore it is possible that all three sub-species in the large derived clade indicated by phylogenetic analyses for both loci and for snail populations (Figs 2 and 3) are in fact O. h. hupensis. The phylogeny estimated for the worms, in which the lowland populations form a clade distinct from those of the highland populations (SAV, SCB and YEB), is consistent with morphological, host-utilisation, and maturation rate observations that suggested independent lineages or sub-species for the highland and lowland S. japonicum in China[63]. Nevertheless, O. h. hupensis appears paraphyletic with two DLB (and one THP) populations forming separate clades near the base of the tree. Divergence of some Hunan/Hubei (DLB) populations away from those of other Lake and Marshland regions could result from the particularly long history of intensive control efforts[4] in these provinces, with slippage of these clades towards the outgroup owing to long branch attraction coupled with a lack of apomorphies among younger clades[64]. It is also interesting to note that the zoophilic strain[65] from Taiwan is not genetically distinct from those strains capable of infecting humans, although it is one of the two most derived members of the lake and marshland clade.
Hypothesis testing
The lack of concordance between the snail and worm phylogenies found in the present study could result from heterochronous evolution of the host and parasite in response to different palaeo-geographical or climatic environments. In the closely related taxon, Schistosoma mekongi, the radiation of the parasite (dated 2.1–1.0 Ma) was shown to be independent of that of its intermediate host (10–5 Ma). Divergence events among the snails were considered to be a concerted response to the final Indosinian orogeny around 5 Ma, with S. mekongi colonising the snails across its present range much later (early Pleistocene)[31,32]. Using a molecular clock the introduction of O. hupensis across mainland China has been dated to the early Miocene (c.a., 22 Ma), with high rates of cladogenesis 8–2 Ma and linked to the exceptionally warm and humid climate in the region at that time and tectonic upheaval in Japan[30]. The divergence of the Schistosoma japonicum clade has been dated at 4.6 Ma[32]; this, implies that the radiation of O. hupensis occurred before that of S. japonicum. If the radiation of the snails and worms is heterochronous there is no opportunity for coevolution; the implication is also that ancestral intermediate hosts differed from those of the present, which again makes coevolution unlikely.
Coevolution might be expected in Schistosoma species such as S. mansoni, which infect snail populations at relatively high prevalence and achieve high rates of cercariogenesis[66], but seems unlikely in S. japonicum because of its lower prevalence in the intermediate host populations. The prevalence of natural infections in China ranges from 0.038% (Jiangsu in 2011) to 7.8% (Anhui in 2013)[67]. In cases where the snails experience a low probability of becoming infected, they are under little pressure to invest resources in defence[68]. In contrast, prevalences as high as 75.7% have been reported for natural populations of S. mansoni in Biomphalaria glabrata in Brazil[69]. Factors such as the generalised nature of gastropod immune systems, and evidence for frequent host switches in the parasites’ evolutionary histories, also make a “Red Queen” scenario unlikely for these schistosomes[32,68,70,71]. Consequently, molecular clock dating for other members of the S. sinensium clade and their intermediate hosts (also close relatives of Oncomelania) and the low prevalence of infection in O. hupensis suggest that the lack of evidence for coevolution found in this study is to be expected.
It could be argued that comparison of naturally infected (and infective) snails, rather than snails from the local populations transmitting the parasite (but not necessarily themselves infected) would be more likely to reveal signs of coevolution, i.e., there could be sub-populations or sub-strains of snails that have been coevolving with the worms, rather than the general snail populations sampled in this study. The existence of such sub-populations is questionable, as is the existence of resistant and susceptible lineages of snails in schistosomiasis. It has been shown that any snail taken from a natural population of B. glabrata will become infected if exposed to enough miracidia from a natural S. mansoni population[72]. Consequently, a “resistant” snail line is merely a sub-population selected to be discordant with the epitopes expressed by a particular Schistosoma line. Even authors working on the B. glabrata-S. mansoni association, have observed a complete reversal in resistance phenotype after a few laboratory generations and note that genotypic responses would only occur in associations where prevalences are high[36]. In view of this, it is unlikely that resistant sub-strains of O. hupensis exist and infection probably occurs more at random across the general snail population (perhaps influenced by ecology and spatial coincidence). Nevertheless, it would be interesting to repeat the present study using naturally infected snails from the localities studied and seek to detect any signs of coevolution.
Phylogeography
As mentioned above, earlier studies date the introduction of O. hupensis to mainland China at around 22 Ma, at which time the region was significantly less mountainous, and three general clades appear to have been rapidly established; these span China, with the O. h. robertsoni clade in the far West, the Dongting Lake Basin (DLB) clade in the center of the Lake and Marshland region, and the Tai Hu Plain (THP) clade near the East coast of mainland China (Fig 6A). O. h. robertsoni appears to have diverged little since its initial colonisation of the mountain valleys in which it is found today, and its lineages have probably been isolated therein since the second major uplift of the Himalaya about 7 Ma[73]. The other two clades appear to have undergone two successive, more recent, cladogenic events, which in the case of the DLB clade, gave rise to the GCP and PLB populations (to the West and East, respectively; Fig 6A node 3). The THP clade gave rise to FCP, Taiwan, Japanese and Philippine populations of O. h. hupensis (Fig 6A node 2). Such a recolonisation of Japan by mainland Chinese O. hupensis is consistent with the “East to West” hypothesis[29]. The divergence of the western clades is likely to have occurred around 8 Ma when the Himalayan uplift altered global climate and triggered increasing aridity in the region, which would have fragmented existing Oncomelania populations[30]. Interestingly, the Taiwan population is also included in the lake and marshland clades even though Taiwan has been separated from the mainland since Pleistocene[74,75]and the more recent divergences within O. hupensis occurred before 2 Ma. It is possible that tsunami events could have exchanged snails between the mainland and Taiwan. Indeed, the March 2011 Pacific tsunami demonstrated that large aggregates of material may cross even oceanic distances in less than 15 months and that freshwater pools on these may harbour viable communities of exotic aquatic organisms (including molluscs)[76]. Oncomelania is also capable of aestivating out of water for several months. In addition, intermittent land bridges occurred, linking Taiwan during the Quaternary[77]. The relative lack of genetic variation among the Taiwan populations also suggests a recent colonisation of the island (or extinction of long established lineages followed by recent recolonisation). The Guangxi Plain (GCP) populations formed a clade distinct from other O. hupensis and may have been isolated from the other O. hupensis taxa since the late Miocene/late Pliocene by uplift along the margins of the Youjiang Basin (Jiangnan range)[78,79].
Although the radiation of S. japonicum is described above as occurring around 4 Ma later than that of the snails, the western (mountain) clades of the parasite still show the same initial divergence and then absence of cladogenesis as do the snails. The ancestral hosts of S. sinensium group parasites appear to be rodents (especially Rattus and its sister group) and it is therefore likely that S. japonicum radiated in China in concert with the Pliocene radiation of Rattus which began in Southeast Asia[80]. After colonising the valleys of Sichuan and Yunnan in rodents radiating into China from Southeast Asia, the parasites would become isolated in these valleys by cooling and increasing aridity in the Pleistocene[81]; thus suppressing further cladogenesis. In contrast, the lake and marshland clades undergo a series of cladogenic events spread from around the early Pliocene towards the Recent. Initially DLB and THP clades are established, together with a second THP clade that is derived from the O. h. robertsoni clades (Fig 6B node 1), this is followed by progressive diversification of one branch of the second THP clade (Fig 6B node 2), whilst the DLB-associated THP clade remains stable. The second THP clade most recently gives rise to an ancestral form that diversifies into a PLB and a Taiwan clade (Fig 6B node 3). The possibility of a long-distance dispersal from Sichuan in the western mountains to THP near the East coast of China (Fig 6B arrow) is an intriguing one; however, the possibility of misidentification or laboratory error concerning Genbank deposited sequences must also be considered where highly inconsistent relationships are found The introduction needs to be dated in future work and might be related to traffic down the Yangtze river (human activities) and the extensive cladogenesis and spread of the Sichuan strain in the coastal lowland areas is an unexpected event, which might relate to better development of the parasite in naïve human hosts (or presence of a more dynamic host population than in the mountain regions).
Analysis of the extended data set (S1 Fig) shows the Japanese S. japonicum arising from the same basal THP lineage as the Taiwan population. In contrast the Philippine clade arises from the basal YEB clade along with taxa from Sichuan. Although this relationship appears to mirror that of the snail phylogeny, it is a relatively recent divergence in the parasites and a relatively earlier one in the snails; thus it does not increase the degree of phylogenetic congruence between the hosts and parasites.
The possibility of extensive radiation and dispersal, after long-distance introduction of a Sichuan strain of S. japonicum to the coastal region, is important in view of the fact that the mountains of Yunnan and Sichuan appear to have formed a barrier to dispersal of O. h. robertsoni transmitted S. japonicum for perhaps millions of years. The observation is particularly relevant in the context of the South-to-North-Water-Transfer project (Eastern Route) which will transfer water from endemic areas in Jiangsu province to areas in Shandong Province, towards the Yellow river, where O. hupensis has yet to be found but where conditions appear to be favorable for transmission[82]. Oncomelania (and the associated schistosomiasis) are most widespread in the lake and marshland areas of the middle and lower Yangtze river drainage; the distribution of snail and parasite in the mountainous areas of Sichuan in more patchy, and in Yunnan they are found only in a restricted area around Dali[83]. As the GMS road projects will breach the mountain ranges between Sichuan and eastern China and Sichuan and Yunnan (and further South into Laos and Thailand), it is important to understand the colonisation history of S. japonicum.
Conclusions
The present work has led to the rejection of the hypothesis of coevolution for Schistosoma japonicum and Oncomelania hupensis on the basis of the samples available. The finding is consistent with models regarding the relative timing of the radiations of the two groups proposed in earlier studies, and with observations of a low prevalence of infection in these snails. Nevertheless, it is still possible that the parasites show some adaptations to a snail population through which they have been cycling for some time, but the analysis does suggest that the worms are not highly evolved/restricted to a particular sub-species or strain of snail. Consequently, host-switching or acquisition is more likely than would be implied by a Red Queen scenario. The findings also suggest that O. h. formosana (of Taiwan) and O. h. tangi (of Fujian) might be O. h. hupensis and not distinct sub-species. The phylogeographical reconstructions suggest that at least one long-distance dispersal event occurred across China between the western mountain populations of S. japonicum and the East coast region. The event appears to have triggered extensive cladogenesis and dispersal (including to Taiwan) on the East coast. Consequently, further work is necessary to confirm and to date this long-distance dispersal and to detect any further such events, so that their origins and driving forces can be determined. Further work is also required with a richer data set as support for some of the clades indicated in the analyses was less than 90%. The findings are particularly important in view of the infrastructure development plans which will breach the mountain barriers between Sichuan, Yunnan and Southeast Asia. The results also have implications for the spread of S. japonicum as, in the absence of coevolution, the parasite may more readily colonise new snail populations to which it is not locally adapted, or even new intermediate host species, and this can facilitate its dispersal into new areas. The work also lends support to the East-West hypothesis for the origin and dispersal of Oncomelania and the Pomatiopsidae.
Supporting Information
File A, Oncomelania populations; File B, Schistosoma populations; File C, extended set of Schistosoma populations.
(ZIP)
Plotted using Marble Virtual Globe (open source). For an interactive projection please use the kml file File C in S1 Dataset also provided in the Supporting Information.
(PNG)
Acknowledgments
Thanks are due to: the Natural History Museum (London) for providing access to library facilities, the Department of Biomedical Informatics and the Ohio State University Medical Center for providing computing resources. In addition, much of the sequence data were produced by many individuals and we thank them for sharing their data via the National Institutes of Health’s Genbank.
Data Availability
All DNA sequence data were taken from the GenBank, or were deposited by the authors therein (accession numbers KR002673-KR002677).
Funding Statement
The authors received no specific funding for this work.
References
- 1. Mcgarvey ST, Carabin H, Balolong E, Belisle P, Fernandez T, Joseph L, et al. Cross-sectional associations between intensity of animal and human infection with Schistosoma japonicum in Western Samar province, Philippines. Bull World Health Organ. 2006;84: 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou XN, Wang L-Y, Chen M-G, Wu X-H, Jiang Q-W, Chen X-Y, et al. The public health significance and control of schistosomiasis in China-then and now. Acta Trop. 2005;96: 97–105. [DOI] [PubMed] [Google Scholar]
- 3. Rollinson D, Southgate VR. The genus Schistosoma: a taxonomic appraisal In: Rollinson D, Simpson AJ., editors. The Biology of Schistosomes: From Genes to Latrines. London: Academic Press; 1987. pp. 1–49. [Google Scholar]
- 4. Ross AG., Sleigh AC, Li YS, Davis GM, Williams GM, Jiang Z, et al. Schistosomiasis in the People’s Republic of China: Prospects and Challenges for the 21st Century. Clin Microbiol Rev. 2001;14: 270–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi F, Zhang Y, Ye P, Lin J, Cai Y, Shen W, et al. Laboratory and field evaluation of Schistosoma japonicum DNA vaccines in sheep and water buffalo in China. Vaccine. 2001;20: 462–467. [DOI] [PubMed] [Google Scholar]
- 6. Liang S, Yang C, Zhong B, Qiu D-C. Re-emerging schistosomiasis in hilly and mountainous areas of Sichuan, China. Bull World Health Organ. 2006;84: 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou Y-B, Yang Mei-Xia, Zhao Gen-Ming, Wei Jiang-Guo, Jiang Qing-Wu. Oncomelania hupensis (Gastropoda: Rissooidea), Intermediate Host of Schistosoma japonicum In China: Genetics and Molecular Phylogeny Based On Amplified Fragment Length Polymorphisms. Malacologia. 2007;49: 367–382. [Google Scholar]
- 8. Wilke T, Davis GM, Qiu D-C, Spear RC. Extreme mitochondrial sequence diversity in the intermediate schistosomiasis host Oncomelania hupensis robertsoni: another case of ancestral polymorphism? Malacologia. 2006;48: 143–157. [Google Scholar]
- 9. Davis GM, Yi Z, Hua GY, Spolsky C. Population genetics and systematic status of Oncomelania hupensis (Gastropoda: Pomatiopsidae) throughout China. Malacologia. 1995;37: 133–156. [Google Scholar]
- 10. Guan F, Niu A, Han Q-X, Chen Q-R. Extended study on CO I sequence variation of Oncomelania hupensis . Int J Med Parasit Dis. 2011;38: 23–27. [Google Scholar]
- 11. Wilke T, Davis GM, Chen C-E, Zhou XN, Zeng XP, Zhang Y, et al. Oncomelania hupensis (Gastropoda: Rissooidea) in eastern China: molecular phylogeny, population structure, and ecology. Acta Trop. 2000;77: 215–227. [DOI] [PubMed] [Google Scholar]
- 12. Shi C-H, Wilke T, Davis GM, Xia M-Y, Qiu C-P. Population genetics, micro-phylogeography, ecology, and susceptibility to schistosome infection of Chinese Oncomelania hupensis hupensis (Gastropoda: Rissooidea: Pomatiopsidae) in the Miao river system. Malacologia. 2002;44: 333–347. [Google Scholar]
- 13. Zhao Q-P, Zhang SH, Deng Z-R, Jiang M-S, Nie P. Conservation and variation in mitochondrial genomes of gastropods Oncomelania hupensis and Tricula hortensis, intermediate host snails of Schistosoma in China. Mol Phylogenet Evol. 2010;57: 215–226. 10.1016/j.ympev.2010.05.026 [DOI] [PubMed] [Google Scholar]
- 14. Li S-Z, Wang Y-X, Liu Q, Lv S, Wang Q, Wu Y, et al. Complete mitochondrial genome sequence of Oncomelania hupensis (Gastropoda: Rissooidea). Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2009;27: 291–296. [PubMed] [Google Scholar]
- 15. Wilke T, Davis GM, Gong X, Liu H-X. Erhaia (Gastropoda: Rissooidea): Phylogenetic relationships and the question of Paragonimus coevolution in Asia. Am J Trop Med Hyg. 2000;62: 453–459. [DOI] [PubMed] [Google Scholar]
- 16. Attwood SW, Ambu S, Meng XH, Upatham ES, Xu F-S, Southgate VR. The phylogenetics of triculine snails (Rissooidea: Pomatiopsidae) from south-east Asia and southern China: historical biogeography and the transmission of human schistosomiasis. J Molluscan Stud. 2003;69: 263–271. [Google Scholar]
- 17. Guan F, Niu AO, Attwood SW, Li YL, Zhang B, Zhu YH. Molecular phylogenetics of Triculine snails (Gastropoda: Pomatiopsidae) from southern China. Mol Phylogenet Evol. 2008;48: 702–707. 10.1016/j.ympev.2008.04.021 [DOI] [PubMed] [Google Scholar]
- 18. Kameda Y, Kato M. Terrestrial invasion of pomatiopsid gastropods in the heavy-snow region of the Japanese Archipelago. BMC Evol Biol. 2011;11: 118 10.1186/1471-2148-11-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li S-Z, Wang Y-X, Yang K, Liu Q, Wang Q, Zhang Y, et al. Landscape genetics: the correlation of spatial and genetic distances of Oncomelania hupensis, the intermediate host snail of Schistosoma japonicum in mainland China. Geospat Health. 2009;3: 221–231. [DOI] [PubMed] [Google Scholar]
- 20. Zhao G-H, Li J, Song H-Q, Li X-Y, Chen F, Lin R-Q, et al. A specific PCR assay for the identification and differentiation of Schistosoma japonicum geographical isolates in mainland China based on analysis of mitochondrial genome sequences. Infect Genet Evol. 2012;12: 1027–1036. 10.1016/j.meegid.2012.02.020 [DOI] [PubMed] [Google Scholar]
- 21. Le TH, Blair D, Agatsuma T, Iwagami M, Humair P-F, Campbell NJ., et al. Phylogenies inferred from mitochondrial gene orders—a cautionary tale from the parasitic flatworms. Mol Biol Evol. 2000;17: 1123–1125. [DOI] [PubMed] [Google Scholar]
- 22. Sorensen E, Drew AC, Brindley PJ, Bogh HO, Gasser RB, Qian BZ, et al. Variation in the sequence of a mitochondrial NADH dehydrogenase I gene fragment among six natural populations of Schistosoma japonicum from China. Int J Parasitol. 1998;28: 1931–1934. [DOI] [PubMed] [Google Scholar]
- 23. Sorensen E, Bogh HO, Johansen MV, McManus DP. PCR-based identification of individuals of Schistosoma japonicum representing different subpopulations using a genetic marker in mitochondrial DNA. Int J Parasitol. 1999;29: 1121–1128. [DOI] [PubMed] [Google Scholar]
- 24. Shrivastava J, Bao ZQ, McVean G, Webster JP. An insight into the genetic variation of Schistosoma japonicum in mainland China using DNA microsatellite markers. Mol Ecol. 2005;14: 839–849. [DOI] [PubMed] [Google Scholar]
- 25. Zhao GH, Mo XH, Zou FC, Li J, Weng YB, Lin RQ, et al. Genetic variability among Schistosoma japonicum isolates from different endemic regions in China revealed by sequences of three mitochondrial DNA genes. Vet Parasitol. 2009;162: 67–74. 10.1016/j.vetpar.2009.02.022 [DOI] [PubMed] [Google Scholar]
- 26. Zhao QP, Jiang MS, Dong HF, Nie P. Diversification of Schistosoma japonicum in Mainland China Revealed by Mitochondrial DNA. PLoS Negl Trop Dis. 2012;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang L, Liu Y, Liao J, Gong P. Wetlands explain most in the genetic divergence pattern of Oncomelania hupensis . Infect Genet Evol. 2014;27: 436–444. 10.1016/j.meegid.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 28. Davis GM. The origin and evolution of the gastropod family Pomatiopsidae, with emphasis on the Mekong river Triculinae. Acad Nat Sci Philad, Monogr. 1979;20: 1–120. [Google Scholar]
- 29. Attwood SW. Mekong Schistosomiasis: Where Did It Come from and Where Is It Going? In: Campbell IC, editor. The Mekong: Biophysical Environment of an International River Basin. New York: Academic Press; 2009. p. 464. [Google Scholar]
- 30. Liu L, Huo G-N, He H-B, Zhou B, Attwood SW. A phylogeny for the pomatiopsidae (Gastropoda: Rissooidea): a resource for taxonomic, parasitological and biodiversity studies. BMC Evolutionary Biology. 2014;14: 29 10.1186/1471-2148-14-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Attwood SW, Fatih FA, Campbell IC, Upatham ES. The distribution of Mekong schistosomiasis, past and future: preliminary indications from an analysis of genetic variation in the intermediate host. Parasitol Int. 2008;57: 256–270. 10.1016/j.parint.2008.04.003 [DOI] [PubMed] [Google Scholar]
- 32. Attwood SW, Fatih FA, Upatham ES. A study of DNA-sequence variation among Schistosoma mekongi (Trematoda: Digenea) populations and related taxa; phylogeography and the current distribution of Asian schistosomiasis. PLoS Negl Trop Dis. 2008;2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davis GM. Snail hosts of Asian Schistosoma infecting man: evolution and coevolution. In: Bruce JI, Sornmani S, Asch HL, Crawford KA, editors. The Mekong Schistosome. Malacological Review, suppl. 2; 1980. pp. 195–238. [Google Scholar]
- 34. Davis GM, Wilke T, Spolsky C, Zhang Y, Xia M- Y, Rosenberg G. Cytochrome Oxidase I-based phylogenetic relationships among the Hydrobiidae, Pomatiopsidae, Rissoidae, and Truncatellidae (Gastropoda: Prosobranchia: Rissoacea). Malacologia. 1998;40: 251–266. [Google Scholar]
- 35. Lockyer AE, Jones CS, Noble LR, Rollinson D. Trematodes and snails: an intimate association. Can J Zool. 2004;82: 251–269. [Google Scholar]
- 36. Webster JP, Shrivastava J, Johnson PJ, Blair L. Is host-schistosome coevolution going anywhere? BMC Evol Biol. 2007;7: 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Webster JP, Davies CM. Coevolution and compatibility in the snail-schistosome system. Parasitology. 2001;123: S41–S56. [DOI] [PubMed] [Google Scholar]
- 38. Van Valen L. A new evolutionary law. Evol Theor. 1973;1: 1–30. [Google Scholar]
- 39. Ishida M. Effectiveness and Challenges of Three Economic Corridors of the Greater Mekong Sub-region. IDE Discussion Papers. 2005;35: 1–22. [Google Scholar]
- 40. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3: 294–299. [PubMed] [Google Scholar]
- 41. Palumbi SR, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The simple fool’s guide to PCR. Version 2.0 Honolulu: University of Hawaii; 1991. [Google Scholar]
- 42. Attwood SW, Brown DS, Meng XH, Southgate VR. A new species of Tricula (Pomatiopsidae: Triculinae) from Sichuan Province, PR China: intermediate host of Schistosoma sinensium . Systematics and Biodiversity. 2003;1: 109–116. [Google Scholar]
- 43. Zarowiecki MZ, Huyse T, Littlewood DT. Making the most of mitochondrial genomes—Markers for phylogeny, molecular ecology and barcodes in Schistosoma (Platyhelmonthes: Digenea). Int J Parasitol. 2007;37: 1401–1418. [DOI] [PubMed] [Google Scholar]
- 44. Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25: 1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27: 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 47. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 49. Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 50. Varón A, Vinh LS, Wheeler WC. POY version 4: phylogenetic analysis using dynamic homologies. Cladistics. 2010;26: 72–85. [DOI] [PubMed] [Google Scholar]
- 51. Wheeler W. Fixed Character States and the Optimization of Molecular Sequence Data. Cladistics. 1999;15: 379–385. [DOI] [PubMed] [Google Scholar]
- 52. Liu L, Mondal M, Idris M, Lokman H, Rajapakse PJ, Satrija F, et al. The phylogeography of Indoplanorbis exustus (Gastropoda: Planorbidae) in Asia. Parasit Vectors. 2010;3: 57 10.1186/1756-3305-3-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Farris JS, Kallersjo M, Kluge AG, Bult C. Constructing a significance test for incongruence. Syst Biol. 1995;44: 570–572. [Google Scholar]
- 54. Swofford D. PAUP: Phylogenetic Analysis Using Parsimony (and other methods). Version 4.0 beta version Sunderland, Massachusetts: Sinauer Associates; 2002. [Google Scholar]
- 55. Lee MS. Uninformative Characters and Apparent Conflict Between Molecules and Morphology. Mol Biol Evol. 2001;18: 676–680. [DOI] [PubMed] [Google Scholar]
- 56. Barker FK, Lutzoni FM. The utility of the incongruence length difference test. Syst Biol. 2002;51: 625–637. [DOI] [PubMed] [Google Scholar]
- 57. Darlu P, Lecointre G. When does the incongruence length difference test fail? Mol Biol Evol. 2002;19: 432–437. [DOI] [PubMed] [Google Scholar]
- 58. Dolphin K, Belshaw R, Orme CD, Quicke DL. Noise and incongruence: interpreting results of the incongruence length difference test. Mol Phylogenet Evol. 2000;17: 401–406. [DOI] [PubMed] [Google Scholar]
- 59. Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic inference In: Hillis DM, Moritz C, Mable B, editors. Molecular Systematics. Sunderland, Massachusetts: Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- 60. Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13: 235–238. [DOI] [PubMed] [Google Scholar]
- 61. Nye TMW, Liò P, Gilks WR. A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics. 2005; 10.1093/bioinformatics/bti720 [DOI] [PubMed] [Google Scholar]
- 62. Wägele JW, Mayer C. Visualizing differences in phylogenetic information content of alignments and distinction of three classes of long-branch effects. BMC Evol Biol. 2007;7: 147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. He YX, Hu YQ, Yu QF, Ni CH, Xue HC, Qiu LS, et al. Strain complex of Schistosoma japonicum in the mainland of China. S E Asian J Trop Med Publ Hlth. 1994;25: 232–242. [PubMed] [Google Scholar]
- 64. Kück P, Mayer C, Wägele J-W, Misof B. Long Branch Effects Distort Maximum Likelihood Phylogenies in Simulations Despite Selection of the Correct Model. PLoS One. 2012;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lo CT, Lee KM. Schistosoma japonicum, zoophilic strain, in Oncomelania hupensis chiui and O. h. formosana: miracidial penetration and comparative histology. J Parasitol. 1995;81: 708–713. [PubMed] [Google Scholar]
- 66. Webster JP, Gower CM, Blair L. Do hosts and parasites coevolve? Empirical support from the Schistosoma system. Am Nat. 2004;164: S33–S51. [DOI] [PubMed] [Google Scholar]
- 67. Berger S. Schistosoma Japonicum: Global Status. Los Angeles: GIDEON Informatics Inc; 2014. [Google Scholar]
- 68. Blair D, Davis GM, Wu B. Evolutionary relationships between trematodes and snails emphasizing schistosomes and paragonimids. Parasitology. 2001;123: S229–S243. [DOI] [PubMed] [Google Scholar]
- 69. Barboza DM, Zhang C, Santos NC, Silva MMBL, Rollemberg CVV, de Amorim FJR, et al. Biomphalaria species distribution and its effect on human Schistosoma mansoni infection in an irrigated area used for rice cultivation in northeast Brazil. Geospat Health. 2012;6: S103–109. [DOI] [PubMed] [Google Scholar]
- 70. Agatsuma T, Iwagami M, Liu CX, Rajapakse RPV., Mondal MM., Kitikoon V, et al. Affinities between Asian non-human Schistosoma species, the S. indicum group, and the African human schistosomes. J Helminthol. 2002;76: 7–19. [DOI] [PubMed] [Google Scholar]
- 71. Attwood SW, Upatham ES, Meng XH, Qiu D-C, Southgate VR. The phylogeography of Asian Schistosoma (Trematoda: Schistosomatidae). Parasitology. 2002;125: 1–13. [DOI] [PubMed] [Google Scholar]
- 72. Mitta G, Adema CM, Gourbal B, Loker ES, Theron A. Compatibility polymorphism in snail/schistosome interactions: From field to theory to molecular mechanisms. Dev Comp Immunol. 2012;37: 1–8. 10.1016/j.dci.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Amano K, Taira A. Two-phase uplift of Higher Himalayas since 17 Ma. Geology. 1992;20: 391–394. [Google Scholar]
- 74. Kizaki K, Oshiro I. Paleogeography of the Rytkyu Islands. Mar Sci Month. 1977;9: 542–549. [Google Scholar]
- 75. Ujiie H, Nakamura T. Temporary change of flowing route of the Kuroshio Current into the Ryukyu Trough since the latest glacier period. Chik Month. 1996;18: 524–530. [Google Scholar]
- 76. Bagulayan A, Bartlett-Roa J, Carter A, Inman B, Keen E, Orenstein E, et al. Journey to the Center of the Gyre: The Fate of the Tohoku Tsunami Debris Field. Oceanography. 2012;25: 200–207. [Google Scholar]
- 77. Qiu Y-X, Fu C-X, Comes HP. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet & Evol. 2011;59: 225–244. [DOI] [PubMed] [Google Scholar]
- 78.Zhou W-B. Structural Characteristics of the Xidamingshan Uplift in Guangxi and Its Relationship with the Adjacent Basins–Basic Science Paper. Masters Thesis, Chinese Geology University. 2005.
- 79. Hou J-J, Liu X-D, You X-Z, Qin H-B. The characteristics of surface deformation grouping of the active strike slip fault system and earthquake activity in western Guangxi, China. Acta Seismol Sin. 1993;6: 549–553. [Google Scholar]
- 80. Chaimanee Y, Jaeger J. Evolution of Rattus (mammalia, Rodentia) during the plio‐pleistocene in Thailand. Hist Biol. 2001;15: 181–191. [Google Scholar]
- 81. Sun Y, An Z. Late Pliocene-Pleistocene changes in mass accumulation rates of eolian deposits on the central Chinese Loess Plateau. J Geophys Res. 2005;110: D23101. [Google Scholar]
- 82. Li Z-Q, Teng W-P, Yu S-X, Han X-J. A Change of Climate Conditions for Growth of Oncomelania and Schistosome. Adv Clim Change Res. 2007;106: 106–110. [Google Scholar]
- 83. Davis GM, Wilke T, Zhang Y, Xu X-J, Qiu C-P, Spolsky C, et al. Snail-Schistosoma, Paragonimus interactions in China: population ecology, genetic diversity, coevolution and emerging diseases. Malacologia. 1999;41: 355–377. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File A, Oncomelania populations; File B, Schistosoma populations; File C, extended set of Schistosoma populations.
(ZIP)
Plotted using Marble Virtual Globe (open source). For an interactive projection please use the kml file File C in S1 Dataset also provided in the Supporting Information.
(PNG)
Data Availability Statement
All DNA sequence data were taken from the GenBank, or were deposited by the authors therein (accession numbers KR002673-KR002677).