Figure 4.
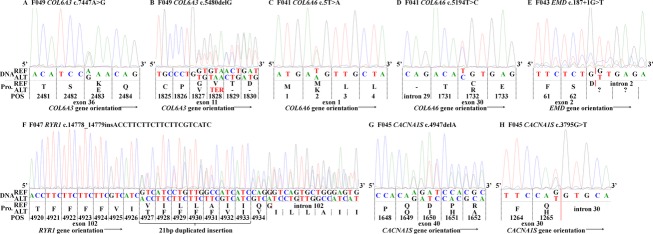
Sanger sequencing traces examples of each pathogenic variant reported from GeneDX clinical confirmation. DNA reference (REF) based on GRCh37. (A) F049 affected male child heterozygous COL6A3 c.7447A>G recessive missense p.Lys2483Glu pathogenic variant. This trace is also similar to the results found in the F041 affected proband (data not shown). (B) F049 affected male child heterozygous COL6A3 c.5480delG recessive frameshift p.Gly1827Valfs*1 pathogenic variant. (C) F041 affected female child heterozygous COL6A6 c.5T>A recessive missense p.Met2Lys likely pathogenic variant. (D) F041 affected female child heterozygous COL6A6 c.5194T>C recessive missense p.Cys1732Arg likely pathogenic variant. (E) F043 affected male child hemizygous EMD c.187+1G>T recessive pathogenic variant. (F) F047 affected male child heterozygous RYR1 c.14778_14779insACCTTCTTCTTCTTCGTCATC dominant duplicated insertion p.Ile4926ins7 (TFFFFVI) pathogenic variant. (G) F045 affected male child heterozygous CACNA1S c.4947delA recessive frameshift p.Gln1649Glnfs*72 pathogenic variant. (H) F045 affected male child heterozygous CACNA1S c.3795G>T recessive missense p.Gln1265His pathogenic variant. Transcripts used for protein position (Pro. POS) for: Col6α3 = NM_004369; Col6α6 = NM_001102608; Emerin = NM_000117; RYR-1 = NM_000540; Cav1.1 = NM_000069. Red solid vertical bars represent splice sites. Dashed black vertical bars represent triplet codon reading frame.
