Abstract
Background
Evidence suggests that early life infections, presence of older siblings and furred pets in the household affect the risk of developing allergic diseases through altered microbial exposure. Recently, low gut microbial diversity during infancy has also been linked with later development of allergies. We investigated whether presence of older siblings, furred pets and early life infections affected gut microbial communities at 9 and 18 months of age and whether these differences were associated with the cumulative prevalence of atopic symptoms of eczema and asthmatic bronchitis at 3 years of age. Bacterial compositions and diversity indices were determined in fecal samples collected from 114 infants in the SKOT I cohort at age 9 and 18 months by 16S rRNA gene sequencing. These were compared to the presence of older siblings, furred pets and early life infections and the cumulative prevalence of diagnosed asthmatic bronchitis and self-reported eczema at 3 years of age.
Results
The number of older siblings correlated positively with bacterial diversity (p = 0.030), diversity of the phyla Firmicutes (p = 0.013) and Bacteroidetes (p = 0.004) and bacterial richness (p = 0.006) at 18 months. Further, having older siblings was associated with increased relative abundance of several bacterial taxa at both 9 and 18 months of age. Compared to the effect of having siblings, presence of household furred pets and early life infections had less pronounced effects on the gut microbiota. Gut microbiota characteristics were not significantly associated with cumulative occurrence of eczema and asthmatic bronchitis during the first 3 years of life.
Conclusions
Presence of older siblings is associated with increased gut microbial diversity and richness during early childhood, which could contribute to the substantiation of the hygiene hypothesis. However, no associations were found between gut microbiota and atopic symptoms of eczema and asthmatic bronchitis during early childhood and thus further studies are required to elucidate whether sibling-associated gut microbial changes influence development of allergies later in childhood.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0477-6) contains supplementary material, which is available to authorized users.
Keywords: Alpha diversity, Asthmatic bronchitis, Early life infections, Eczema furred pets, Infants, Gut microbiota, Older siblings, Richness, SKOT I cohort
Background
More than two decades ago, David Strachan proposed that the inverse relationship observed between household size and prevalence of hay fever and eczema could be explained if allergic diseases were prevented by infection in early childhood, transmitted by unhygienic contact with older siblings [1]. This would later become known as the hygiene hypothesis. Since then, numerous epidemiological studies have confirmed his results, as reviewed by Karmaus and Botezan [2]. A recent comprehensive worldwide study from the International Study of Asthma and Allergies in Children (ISAAC) concluded that eczema and hay fever are indeed inversely associated with number of older siblings, and that this association is mainly seen in affluent countries [3]. Additional environmental factors affecting the risk of allergies have been identified, such as furred pets, which are associated with decreased risk [4, 5] and birth by cesarean section [6] and use of antibiotics [7], which are both associated with increased risk of allergies. Obviously, these associations are not solely due to transmission of infectious microbes, but rather reflect a generally altered exposure to non-pathogenic microbes [8]. This suggests that not only early life infections, but many aspects of altered microbial exposure are affecting development of allergies. Specifically, the important role of the endogenous gut microbiota and its interaction with the immune system has become evident [9]. In particular a reduced diversity in the early gut microbiota has recently been linked with development of both eczema and asthma [10–14]. However, very limited research has presently been done to address the impact of early life infections, older siblings and furred pets on development of the infant gut microbiota, and thus it is still largely unknown how these are linked to development of allergies. Therefore, we aimed to investigate associations between (i) environmental factors, including older siblings, furred household pets and early life infections, (ii) infant microbial gut communities at ages 9 and 18 months and (iii) the cumulative prevalence of atopic symptoms of eczema and asthmatic bronchitis at the age of 3 years in 114 Danish children within the SKOT I cohort [15].
Methods
The SKOT I cohort
The present study is based on the SKOT I cohort, which includes 311 demographically similar Danish children followed during the first 3 years of life with the overall aim to elucidate relationships between early diet, growth and development and later disease risks [15–17]. Background characteristics of the study population has been published previously [16, 18] and we have previously described the gut bacterial population of this cohort [17], however with a less comprehensive methodology than applied here. The study protocol was approved by the Committees on Biomedical Research Ethics for the Capital Region of Denmark (H-KF-2007-0003) and written consent was given by parents. Inclusion criteria for the SKOT I cohort were single birth and full term delivery, absence of chronic illness and age of 9 months ± 2 weeks at start of study. The participants in the SKOT I cohort were recruited by sending an invitation to a random sample of 2211 families [15]. Fifteen percent responded (330), 19 dropped out before first examination and thus 311 were included in the cohort. 290 of the infants were seen both at the 9 and 18 month visit. After exclusion of individuals with fecal samples taken at only one time point, use of antibiotics and inappropriately stored fecal samples (227), a randomly selected subset of 114 samples was used in the present study. Fecal samples obtained at 9 and 18 months of age were freshly delivered on the morning of visitation or had been stored in the participant’s home, in provided freezer containers, either in the freezer (−18 °C) or in the fridge (4 °C) for maximally 24 h before delivery to University of Copenhagen, Department of Nutrition, Exercise and Sports, where they were stored at −80 °C until DNA extraction. Information about household older siblings (categorized into 0, 1 or >2, due to very few infants having more than 2 older siblings) and furred pets (cats, dogs or rabbits in the household during the first 18 months of life) was collected at parental interviews, which were performed at the university and carried out by a trained staff at 9 and 18 months of age, respectively (Additional file 1: Table S1). Early life infections (defined as parentally reported recurrent otitis media or pneumonia initiated before the age of 12 months), allergic heredity (parents and/or siblings diagnosed with food allergy, eczema, hay fever, urticaria or asthma) and cumulative prevalence (0–3 years of age) of diagnosed asthmatic bronchitis and self-reported eczema were collected at parental interviews at 3 years of age (Additional file 1: Table S1). Possible confounding factors on gut microbiota including mode of delivery, gestational age at birth, infant age at 9 and 18 month examinations, age at start of daycare/nursery, breast feeding duration and daily macronutrient intake at 9 and 18 months of age (Additional file 2: Table S2) were used to evaluate possible differences between infants with or without older siblings, furred pets and early life infections.
DNA extraction and PCR amplification of the 16S rRNA gene
DNA was extracted (PowerLyzer® PowerSoil® DNA isolation kit, MoBio 12855–100) from 250 mg feces according to the provided protocol with minor modifications: bead beating was performed at 30 cycles/s for 10 min (Retsch MM 300 mixer mill) and the initial centrifugation steps were performed at 10,000 × g for 3 min, as recommended for clay matter. DNA quantity and quality were measured by Qubit® dsDNA BR assay (Invitrogen™, Q32850) and NanoDrop® 1000 (Thermo Scientific), respectively, yielding 33.3 ± 22.2 ng/μl DNA with A260/A280 = 1.82 ± 0.12 and A260/A230 = 1.61 ± 0.38. The PCR amplification of the V3-region of the 16S rRNA gene was performed with 5 ng community DNA as template, using 0.2 μl Phusion High-Fidelity DNA polymerase (Fisher Scientific, F-553 L), 4 μl HF-buffer, 0.4 μl dNTP (10 mM of each base), 1 μM forward primer (PBU 5′-A-adapter-TCAG-barcode-CCTACGGGAGGCAGCAG-3′) and 1 μM reverse primer (PBR 5′-trP1-adapter-ATTACCGCGGCTGCTGG-3′) in a 20 μl total reaction volume. Both primers included sequencing adaptors and the forward primer additionally a unique 10–12 bp barcode (Ion Xpress™ Barcode Adapters). The PCR program included 30s at 98 °C, 24 cycles of 15 s at 98 °C and 30 s at 72 °C, followed by 5 min at 72 °C. The PCR product was purified using HighPrep™ PCR Magnetic Beads (MAGBIO®, AC-60005) with the 96-well magnet stand (MAGBIO®, MyMag 96), according to the prescribed procedure. DNA quantity was measured using Qubit® dsDNA HS assay (Invitrogen™, Q32851) and samples were pooled to obtain equimolar libraries containing up to 90 samples in each library.
Sequencing and data handling
Sequencing of the 16S rRNA gene libraries was performed using the Ion OneTouch™ and Ion PGM systems with a 318-Chip, generating 5–7 million reads per chip with a median length of 180 bp. Sequencing data were imported into CLC Genomic Workbench (version 7.0.3, CLC bio, Qiagen, Aarhus, DK), reads were demultiplexed and trimmed to remove low quality sequences (pbase-calling error = 0.05), ambiguous nucleotides (maximally 2 allowed), primers and barcodes and to discard reads below 110 bp and above 180 bp. Sequencing data is deposited at NCBI Sequence Read Archive with the Accession Number SRP052851, under the BioProject Accession Number PRJNA273694. The sorted and trimmed FASTA files were run through the RDP classifier [19] with a bootstrap cutoff of 50 % as recommended for sequences shorter than 250 bp [20]. The total number of reads for each sample was 46418 ± 17806 and was used to calculate the relative abundance of each bacterial group. In the further analysis a cutoff of 0.01 % in mean relative abundance at either 9 or 18 months was applied. Based on the detection limit (1 read), a threshold was set to 0.001 % (~0.46 reads) and zeroes were assigned this value.
Data analysis and statistical tests
Alpha diversity (Shannon index) was calculated at ages 9 and 18 months based on relative abundance of all identified genera (bacterial diversity) or all identified genera within the four major infant gut associated phyla of Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria (phylum diversity) in each sample, using R (version 3.1.0, R Core Team 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL: http://www.R-project.org/) package vegan [21]. Richness was assessed by randomly subsampling 8000 reads per sample and calculating the average number of observed genera within each sample using the R package vegan. Statistical tests were performed in GraphPad Prism (version 5.0.3, GraphPad Software Inc., La Jolla, CA). Non-parametric Mann-Whitney tests were used to test for differences in medians of alpha diversity and richness between children with and without older siblings, furred pets and early life infections and between children with or without asthmatic bronchitis or eczema during the first 3 years of life. Spearman correlation analyses were performed to address associations between older siblings (0, 1 or >2), furred pets, early life infections and the relative abundance of gut bacterial genera at 9 and 18 months of age, which were also correlated with the binary variables asthmatic bronchitis and eczema during the first 3 years of life. Correlation matrices were illustrated using the R package corrplot [22]. P-values were adjusted for multiple testing using a false discovery rate of 10 % [23]. Using the R package FactoMineR [24] principal components were calculated from data on relative abundance of all bacterial families.
Results
Effect of older siblings, furred pets and early life infections on gut microbiota
To minimize confounding effects, we confirmed that prevalence of allergic heredity and C-section, average gestational age at birth, actual age at 9 and 18 month visits, infant age at start of daycare or nursery, breastfeeding duration and macronutrient intake at 9 and 18 months visits were similar between infants with and without older siblings, furred pets or early life infections (Additional file 2: Table S2). Alpha diversity and richness of the gut microbial populations were calculated for all individuals at ages 9 and 18 months, and these data were compared between infants with and without older siblings, furred pets and early life infections (Fig. 1). Infants with older siblings in the household had a higher bacterial diversity (p = 0.045) and richness (p = 0.009) at 18, but not at 9 months of age compared to infants with no older siblings (Fig. 1a, b). In addition, significant correlations between the specific number of older siblings (0, 1 or >2) and bacterial diversity (p = 0.030) or richness (p = 0.006) were found (Fig. 2a, b). Specifically, the phyla diversity within Firmicutes (p = 0.013) and Bacteroidetes at age 18 months (p = 0.004) were positively correlated with numbers of older siblings (Fig. 2c, d), while phyla diversity within Actinobacteria and Proteobacteria were not affected (data not shown). Presence of furred pets in the household did not affect bacterial diversity or richness of the total gut microbial populations at 9 or 18 months (Fig. 1c, d). However, infants with furred pets had lower diversity (p = 0.010) within the Firmicutes phylum at 9 months (data not shown). Infants with registered history of early life infections had a lower bacterial diversity (p = 0.067) and richness (p = 0.023) at age 18 months, but not at age 9 months (Fig. 1e, f). Investigation of associations between abundances of specific microbial genera and the presence of older siblings, furred pets or early life infections (Fig. 3) revealed that Haemophilus and Faecalibacterium abundance at age 9 months were significantly positively associated with the presence of older siblings, while this was true for Barnesiella, Odoribacter, Asaccharobacter and Gondonibacter at age 18 months. The presence of furred pets was positively associated only with Cronobacter abundance at 18 months of age. Early life infections were not significantly associated with any specific gut microbial genera after adjustment for multiple testing. However, we note that Haemophilus abundance at 9 months was positively associated (non-adjusted p = 0.019) with early life infections (Additional file 3: Figure S1).
Fig. 1.
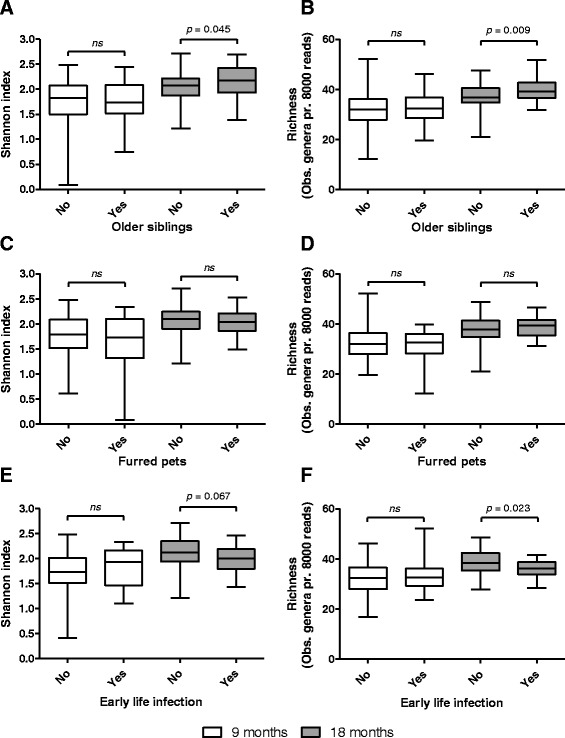
Boxplots comparing bacterial diversity and richness at 9 months (white color) and 18 months (grey color) between individuals with or without older siblings (a, b), furred pets (c, d) and early life infections (e, f). Boxes show 25th to 75th percentiles and whiskers indicate minimum to maximum values. Statistical significance was evaluated by Mann Whitney test, using p < 0.05 as measure of significance. ns = not significant, however p-values below 0.1 are shown
Fig. 2.
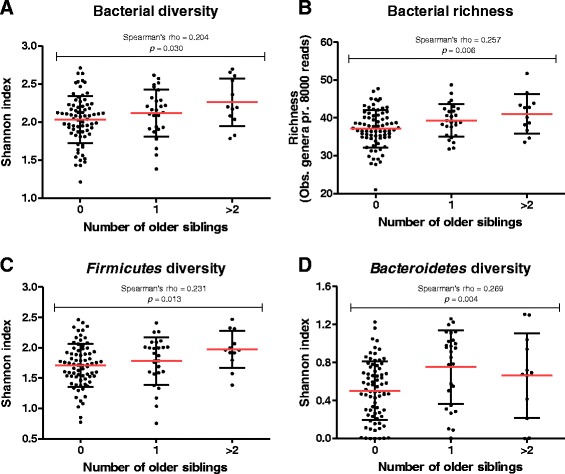
Scatter plots illustrating the bacterial diversity (a), richness (b) and phyla diversities of Firmicutes (c) and Bacteroidetes (d) in infants at 18 months of age divided into groups according to the number of older siblings in the household. Red lines indicate mean ± standard deviation. Spearman’s correlations coefficient rho and p-value (p < 0.05 as a measure of significance) are shown
Fig. 3.
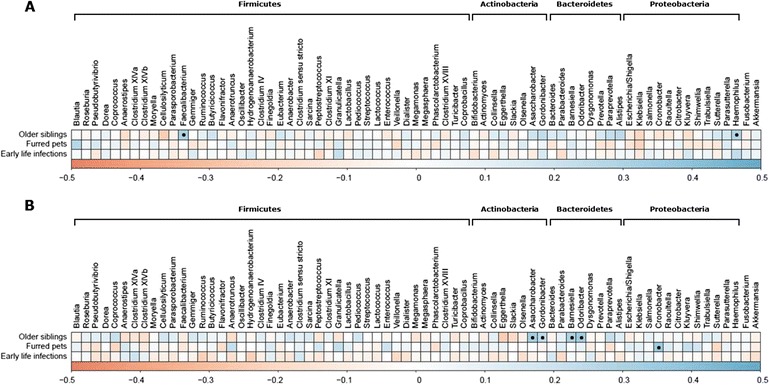
Correlation matrices relating relative abundance of bacterial genera at 9 months (a) and 18 months (b) to the presence of older siblings, furred pets and early life infections. Scale indicate the Spearman’s rank correlation coefficient rho, ranging from −0.5 (negative correlation; red color) to 0.5 (positive correlation; blue color). Black dots indicate statistically significant correlations with FDR adjusted p-value < 0.1
Effect of gut microbiota on cumulative prevalence of eczema and asthmatic bronchitis
Principal component analysis of the gut microbiota composition at family level at ages 9 and 18 months did not reveal any separation of samples originating from children with symptoms of eczema, asthmatic bronchitis or both, compared to samples originating from children without these symptoms (Fig. 4). In accordance with this, Spearman correlation analysis of relative abundances of bacterial genera at 9 and 18 months of age against the occurrence of asthmatic bronchitis and eczema during the first 3 years of life revealed no significant correlations with these outcomes after correction for multiple testing (Additional file 4: Figure S2). Neither asthmatic bronchitis nor eczema was found to be associated with diversity or richness of the faecal microbial populations (Additional file 5: Figure S3).
Fig. 4.
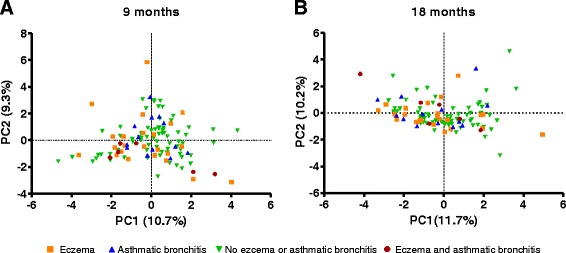
Principle Component Analysis plots of relative abundance of gut bacterial families at 9 (a) and 18 months of age (b). Green triangles indicate no eczema or asthmatic bronchitis, orange squares indicate presence of eczema, blue triangles indicate presence of asthmatic bronchitis and red circles indicate presence of both asthmatic bronchitis and eczema
Discussion
We recently, using less comprehensive methodology, profiled the gut microbiota development of the cohort analysed in the present study [17], yet the possible associations with atopic symptoms and presence of early life infections, furred pets and older siblings in the household were not previously analysed. When assessing the effects of these external factors on infant gut microbiota, gestational age at birth [25], mode of delivery [26], infant age at sampling [27], start of daycare or nursery, breast feeding [17], dietary patterns [28] and use of antibiotics [29] are among the most likely possible confounding factors. We therefore excluded infants with current use of antibiotics and confirmed that C-section prevalence, average gestational age at birth, actual infant age at 9 and 18 month visits, infant age at start of daycare or nursery, breast feeding duration and nutrient intake at 9 or 18 months of age between infants with and without older siblings, furred pets and early life infections were similar (Additional file 2: Table S2). Judged by the high similarity between these groups, these factors seem unlikely to confound our results. However, we cannot exclude the possibility that other factors might affect our results.
In general, it is a limitation of our study that a relatively low number of infants had early life infections and furred pets, which might explain that only few associations were observed. Nonetheless, in children with registered early life infections, diversity and richness measures were lower at 18 months, but not at 9 months than in children with no early life infections. For most subjects’ recurrence of infections were present between 9 and 18 months (Additional file 1: Table S1). Thus, even though we excluded individuals with current antibiotics use at time of sampling and recorded no use of antibiotics 7-days prior to 18 month visits, earlier use in connection with an infection might still impact diversity/richness at the sampling point of 18 months. In agreement with this, antibiotic consumption is reported to reduce bacterial alpha diversity as well as richness, and the effects may be present for a long time after the treatment has ended [30, 31]. Presumably, potential beneficial effects of early life infections on the development of allergies [32] might be counteracted by treatment of these infections with antibiotics [7].
Infants with furred pets had a lower Firmicutes diversity at 9 months of age and higher abundance of Cronobacter at 18 months of age. Very few studies have included investigations of the effects of furred pets on gut microbiota composition [33–35]. While a study in 24 Canadian infants at 4 months of age found over-representation of Clostridiaceae, Veillonella, Peptostreptococcaceae and Coprococcus and under-representation of Bifidobacteriaceae in infants living with pets [34], two studies in much larger cohorts found no effect of having pets in the household at 1 and 6 months on the gut microbiota composition [33, 35]. Obviously, while differences in pet-types and exposure time could explain these discrepancies, better controlled studies are required to assess the impact of different pets on gut microbiota.
The presence of older siblings in the household significantly affected the infant gut microbial community. Bacterial diversity and richness were significantly higher at 18 months, but not at 9 months for infants having one or more older siblings, compared to infants without older siblings. The absence of a sibling-effect on diversity/richness at 9 months is in agreement with a previous study reporting no significant differences in Shannon index and Chao1 richness estimate between infants with and without older siblings at age 4 months [34]. Also the specific number of older siblings was positively associated with bacterial alpha diversity and richness as well as diversity within the Bacteroidetes and Firmicutes phyla that are both typical in an adult-like microbiota composition. We suggest that the effect of siblings on gut microbial diversity/richness increases with time during the first years of life, due to increased contact with older siblings. Additionally, it is likely that parental behaviour, such as hygiene practices, is altered as more children are born in the family. We find it plausible that presence of older siblings in the household is more likely to affect the number of different bacteria to which an infant is exposed, affecting richness and diversity, than to affect the relative abundances of specific bacterial taxa in the gut. However, we did observe that the relative abundance of a few genera, namely Faecalibacterium and Haemophilus at 9 months was higher in infants with older siblings. Faecalibacterium prausnitzii is one of the most abundant species within the gastrointestinal tract of adult humans and its abundance rises quickly during the first years of life [17]. It contributes to butyrate production by degradation of non-digestible dietary fibres like pectin and inulin [36], and has been reported to elicit anti-inflammatory effects in vitro [37]. Haemophilus species colonize the nasopharynx and upper respiratory tract in approximately 60 % of non-symptomatic children [38]. Thus, older siblings might transfer these microbes through close contact. A particularly interesting notion though, was that Haemophilus levels at 9 months were also positively associated with early life infections (otitis media or pneumonia). Haemophilus influenzae is known to be involved in both pneumonia and otitis media [39, 40] and is suggested to be transferred from older siblings with these infections [41] or by other children at day-care facilities [38], which might explain our observations. At age 18 months, strict anaerobic genera within the Bacteroidetes phylum (Odoribacter and Barnesiella) were positively correlated with the presence of older siblings. This is consistent with the increased diversity of Bacteroidetes observed in infants with older siblings. In support of this, a previous study showed that the ratio of strict to facultative anaerobes 12 months after birth was lower in infants without older siblings than infants with older siblings [33], suggesting that infants acquire strict anaerobic bacteria from their older siblings. Only few other studies have addressed the effect of older siblings on specific gut microbes. These studies have reported that having older siblings is associated with increased colonization rates of Lactobacillus and Bacteroides and decreased colonization rates of Clostridium at age 5 weeks [42], lower abundance of Peptostreptococcaceae at age 4 months [34], as well as higher abundance of Bifidobacterium and lower abundance of Enterobacteriaceae during infancy [43]. While we could not confirm these results, difference in age, geography, culture, eating habits and other confounding factors or different methodological procedures could explain that studies report different outcomes with respect to effects on specific bacterial taxa.
Several studies have reported a link between reduced gut microbial diversity in infancy and later development of eczema [10, 11, 13, 14] and asthma [12], whereas another large study involving a high risk population, found no associations between diversity and eczema or asthma [44]. With the given power, we found no association between gut microbial composition and diversity at 9 or 18 months and occurrence of atopic symptoms of eczema or asthmatic bronchitis assessed during the first 3 years of life. However, excluding infants with atopic symptoms of eczema before the age of 18 months (n = 10), we saw a tendency (p = 0.079) for reduced bacterial diversity in infants with atopic symptoms eczema occurring after the age of 18 months (n = 19). This could be relevant since the effect of older siblings on bacterial diversity seems to be evident at 18 months, but not 9 months of age. Most previous studies reporting such an association measured gut microbial diversity in early infancy [10–14, 44], typically the first weeks or months of life, where the gut microbiota is more unstable than in late infancy/early childhood [45, 46], possibly explaining the differences in results. Only one previous study included bacterial diversity at 18 months of age and reported a higher diversity in individuals with eczema compared to healthy controls [47]. However, this was based on only 24 individuals and dietary patterns at 18 months were not recorded, despite that diet is one of the strongest factors affecting gut microbiota [48] and thus a potential confounder. Differences in methods of assessing atopic symptoms could also contribute to inconsistencies between results. The fact that eczema was parentally self-reported is a limitation of our study, since a number of other skin symptoms might be interpreted as eczema [49]. Furthermore, asthmatic bronchitis is often caused by a viral infection in early childhood and might not be indicative of actual asthma development [50]. Recall bias is another issue related to questionnaires assessing symptoms of atopy. Therefore, inadequate assessment of what are actual symptoms of allergy might explain discrepancies compared to previous studies reporting a link between reduced gut microbial diversity and allergy development. In light of the recent strong epidemiological evidence provided by ISAAC [3] of a sibling effect on prevalence of eczema in children at both 6–7 years and 13–14 years of age, we suspect that the increased gut microbial diversity/richness in early childhood, associated with the number of older siblings, could contribute to the lower eczema prevalence observed later in childhood.
Conclusion
We found that the presence of older siblings in the household significantly associates with increased gut microbial diversity and richness during early childhood. This has to our knowledge not previously been shown and could contribute to the substantiation of the hygiene hypothesis. However, gut microbiota during early childhood was not associated with cumulative occurrence of atopic symptoms of eczema or asthmatic bronchitis at 3 years of age. Further studies are warranted to elucidate the possible implications of sibling-associated gut microbial changes on development of allergies in later childhood.
Acknowledgement
We thank the children and families participating in the SKOT I study, which was supported by the Danish Directorate for Food, Fisheries and Agribusiness (Grant no. 3304-FSE-06-0503). The microbiota analysis was supported by the Gut, Grain and Greens (3G) Center financed by the Danish Council for strategic research (Grant no. 11–116163). Bodil Madsen is acknowledged for technical lab support regarding DNA purification, PCR amplification and library preparation.
Abbreviations
- SKOT I
a Danish abbreviation for “Dietary habits and wellbeing of young children”
Additional files
Number of children with early life infections, older siblings, furred pets, family history of allergy and cumulative prevalence of reported eczema and asthmatic bronchitis assessed at 3 years of age.
Possible confounding factors for effects of older siblings, furred pets and early life infections on gut microbiota: Family history of allergy, C-section, gestational age at birth, infant age at 9 and 18 month visits, infant age at start of daycare/nursery, duration of breastfeeding and daily nutrient intake at 9 and 18 months of age for infants with and without older siblings, furred pets and early life infections.
Relative abundance of Haemophilus at 9 and 18 months of age in individuals with or without early life infections (A) and older siblings (B).
Correlation matrices relating relative abundance of gut bacterial genera at 9 months (A) and 18 months (B) to the presence of asthmatic bronchitis and eczema.
Boxplots comparing bacterial diversity and richness at 9 and 18 months between individuals with or without eczema (A-B) and asthmatic bronchitis (C-D).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KFM designed the SKOT I study and was in charge of data collection from the cohort. AH and GZ developed questionnaires for assessment of atopic symptoms. MFL performed fecal sample DNA purification, PCR amplification and library preparation for 16S rRNA gene sequencing. TRL, MFL, AB and MIB conceived of and designed the microbiota analysis, and MFL performed this analysis and drafted the manuscript. All authors participated in interpretation and discussion of results and have read and approved the final manuscript.
Contributor Information
Martin Frederik Laursen, Phone: +45 35887017, Email: mfrla@food.dtu.dk.
Gitte Zachariassen, Email: gitte.zachariassen@rsyd.dk.
Martin Iain Bahl, Email: mbah@food.dtu.dk.
Anders Bergström, Email: bergstrom@nexs.ku.dk.
Arne Høst, Email: Arne.Hoest@rsyd.dk.
Kim F. Michaelsen, Email: kfm@nexs.ku.dk
References
- 1.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–60. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karmaus W, Botezan C. Does a higher number of siblings protect against the development of allergy and asthma? A review. J Epidemiol Community Heal. 2002;56:209–217. doi: 10.1136/jech.56.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strachan DP, Aït-Khaled N, Foliaki S, Mallol J, Odhiambo J, Pearce N, et al. Siblings, Asthma, Rhinoconjunctivitis And Eczema: A Worldwide Perspective From The International Study Of Asthma And Allergies In Childhood. Clin Exp Allergy. 2014;45:126–36. [DOI] [PMC free article] [PubMed]
- 4.Nafstad P, Magnus P, Gaarder PI, Jaakkola JJK. Exposure to pets and atopy-related diseases in the first 4 years of life. Allergy. 2001;56:307–312. doi: 10.1034/j.1398-9995.2001.00881.x. [DOI] [PubMed] [Google Scholar]
- 5.Holscher B, Frye C, Wichmann H-E, Heinrich J. Exposure to pets and allergies in children. Pediatr Allergy Immunol. 2002;13:334–341. doi: 10.1034/j.1399-3038.2002.02063.x. [DOI] [PubMed] [Google Scholar]
- 6.Renz-Polster H, David MR, Buist AS, Vollmer WM, O’Connor EA, Frazier EA, et al. Caesarean section delivery and the risk of allergic disorders in childhood. Clin Exp Allergy. 2005;35:1466–72. [DOI] [PubMed]
- 7.Droste JHJ, Wieringa MH, Weyler JJ, Nelen VJ, Vermeire PA, Van Bever HP. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease? Clin Exp Allergy. 2000;30:1548–1553. doi: 10.1046/j.1365-2222.2000.00939.x. [DOI] [PubMed] [Google Scholar]
- 8.Flohr C, Yeo L. Atopic dermatitis and the hygiene hypothesis revisited. Curr Probl Dermatol. 2011;41:1–34. doi: 10.1159/000323290. [DOI] [PubMed] [Google Scholar]
- 9.Noverr MC, Huffnagle GB. The “microflora hypothesis” of allergic diseases. Clin Exp Allergy. 2005;35:1511–20. doi: 10.1111/j.1365-2222.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 10.Ismail IH, Oppedisano F, Joseph SJ, Boyle RJ, Licciardi PV, Robins-Browne RM, et al. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr Allergy Immunol. 2012;23:674–81. [DOI] [PubMed]
- 11.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434–440. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–50. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129–34. [DOI] [PubMed]
- 14.Forno E, Onderdonk AB, McCracken J, Litonjua AA, Laskey D, Delaney ML, et al. Diversity of the gut microbiota and eczema in early life. Clin Mol Allergy. 2008;6:11. [DOI] [PMC free article] [PubMed]
- 15.Madsen AL, Schack-Nielsen L, Larnkjaer A, Mølgaard C, Michaelsen KF. Determinants of blood glucose and insulin in healthy 9-month-old term Danish infants; the SKOT cohort. Diabet Med. 2010;27:1350–7. doi: 10.1111/j.1464-5491.2010.03134.x. [DOI] [PubMed] [Google Scholar]
- 16.Arnberg K, Østergård M, Madsen AL, Krarup H, Michaelsen KF, Mølgaard C. Associations between vitamin D status in infants and blood lipids, body mass index and waist circumference. Acta Paediatr. 2011;100:1244–8. doi: 10.1111/j.1651-2227.2011.02273.x. [DOI] [PubMed] [Google Scholar]
- 17.Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014;80:2889–900. [DOI] [PMC free article] [PubMed]
- 18.Andersen LBB, Pipper CB, Trolle E, Bro R, Larnkjær A, Carlsen EM, et al. Maternal obesity and offspring dietary patterns at 9 months of age. Eur J Clin Nutr. 2015;69(6):668–75. [DOI] [PubMed]
- 19.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claesson MJ, O’Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. [DOI] [PMC free article] [PubMed]
- 21.Oksanen AJ, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. Vegan: Community Ecology Package. R Packag version 2.0–10 http://CRAN.R-project.org/package=vegan. 2015.
- 22.Package T, Wei AT. Corrplot: Visualization of a correlation matrix. R Packag version 0.73 http://cran.r-project.org/package=corrplot. 2015.
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300.
- 24.Husson AF, Josse J, Le S, Mazet J, Husson MF. FactoMineR: Multivariate Exploratory Data Analysis and Data Mining with R. R Packag version 1.26 http://CRAN.R-project.org/package=FactoMineR. 2015.
- 25.Dogra S, Sakwinska O, Soh S-E, Ngom-Bru C, Brück WM, Berger B, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio 2015, 6. doi:10.1128/mBio.02419-14 [DOI] [PMC free article] [PubMed]
- 26.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–66. [DOI] [PubMed]
- 27.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. [DOI] [PMC free article] [PubMed]
- 28.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed]
- 29.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–75. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108:4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fishbein AB, Fuleihan RL. The hygiene hypothesis revisited: does exposure to infectious agents protect us from allergy? Curr Opin Pediatr. 2012;24:98–102. doi: 10.1097/MOP.0b013e32834ee57c. [DOI] [PubMed] [Google Scholar]
- 33.Adlerberth I, Strachan DP, Matricardi PM, Ahrné S, Orfei L, Aberg N, et al. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol. 2007;120:343–50. [DOI] [PubMed]
- 34.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Sears MR, et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol. 2013;9:15. [DOI] [PMC free article] [PubMed]
- 35.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. [DOI] [PubMed]
- 36.Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJM, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonicFaecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derivedsubstrates for growth. Appl Environ Microbiol. 2012;78:420–8. [DOI] [PMC free article] [PubMed]
- 37.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. [DOI] [PMC free article] [PubMed]
- 38.Farjo RS, Foxman B, Patel MJ, Zhang L, Pettigrew MM, McCoy SI, et al. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr Infect Dis J. 2004;23:41–6. [DOI] [PubMed]
- 39.Rudan I, Boschi-Pinto C, Biloglav Z, Mulholland K, Campbell H. Epidemiology and etiology of childhood pneumonia. Bull World Health Organ. 2008;86:408–416B. doi: 10.2471/BLT.07.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudley S, Ashe K, Winther B, Hendley JO. Bacterial pathogens of otitis media and sinusitis: detection in the nasopharynx with selective agar media. J Lab Clin Med. 2001;138:338–42. doi: 10.1067/mlc.2001.119311. [DOI] [PubMed] [Google Scholar]
- 41.Loos BG, Bernstein JM, Dryja DM, Murphy TF, Dickinson DP. Determination of the epidemiology and transmission of nontypable Haemophilus influenzae in children with otitis media by comparison of total genomic DNA restriction fingerprints. Infect Immun. 1989;57:2751–2757. doi: 10.1128/iai.57.9.2751-2757.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013;132:601–7. [DOI] [PubMed]
- 43.Yap GC, Chee KK, Hong P-Y, Lay C, Satria CD, Sumadiono, et al. Evaluation of stool microbiota signatures in two cohorts of Asian (Singapore and Indonesia) newborns at risk of atopy. BMC Microbiol. 2011;11:193. [DOI] [PMC free article] [PubMed]
- 44.Bisgaard H, Li N, Bonnelykke K, Chawes BLK, Skov T, Paludan-Müller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–52. [DOI] [PubMed]
- 45.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108:4578–85. [DOI] [PMC free article] [PubMed]
- 47.Nylund L, Satokari R, Nikkilä J, Rajilić-Stojanović M, Kalliomäki M, Isolauri E, et al. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol. 2013;13:12. [DOI] [PMC free article] [PubMed]
- 48.Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, et al. Diet Dominates Host Genotype in Shaping the Murine Gut Microbiota. Cell Host Microbe. 2014;17:72–84. [DOI] [PMC free article] [PubMed]
- 49.Jøhnke H, Vach W, Norberg LA, Bindslev-Jensen C, Høst A, Andersen KE. A comparison between criteria for diagnosing atopic eczema in infants. Br J Dermatol. 2005;153:352–8. doi: 10.1111/j.1365-2133.2005.06491.x. [DOI] [PubMed] [Google Scholar]
- 50.Guilbert TW, Mauger DT, Lemanske RF. Childhood asthma-predictive phenotype. J Allergy Clin Immunol Pract. 2014;2:664–70. doi: 10.1016/j.jaip.2014.09.010. [DOI] [PubMed] [Google Scholar]


