Fig. 1. An integrated model for the repeat expansion mechanism in mammals.
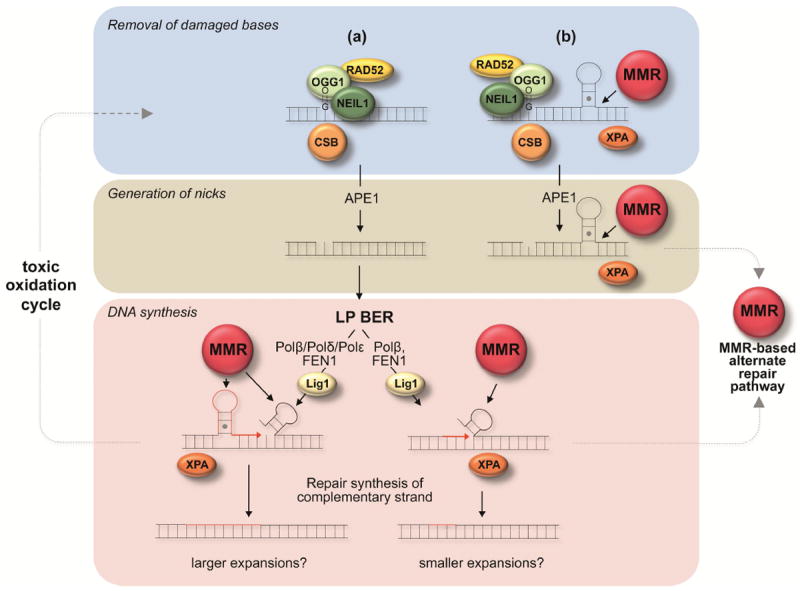
Proteins directly implicated in generating expansions in mouse models or human cells are shown as colored spheres. MMR refers to a complex consisting of the MMR proteins MutSα or MutSβ together with either MutLα or MutLγ. Red strands represent newly synthesized DNA. The repair of DNA damage within the repeat is initiated by DNA glycosylases in response to oxidized bases. This is followed by the removal of the abasic site by APE1 to generate a nick. (a) Repair of the nick may proceed via an LP BER pathway that involves Polβ, Polδ and perhaps Polε [111]. Strand-slippage/hairpin formation at the 3’ terminus of the nascent strand arising during strand displacement synthesis by Polδ/Polε could result in expansion if the hairpin is not removed because Polβ synthesis prevents proof-reading by Polδ/Polε [105]. Formation of a secondary structure on a displaced flap could also result in expansion if proper processing were blocked. A second LP BER pathway that involves Polβ but not Polδ/Polε may generate small expansions by stepwise and distributive gap-filling by Polβ and single-nucleotide gap formation by FEN1 [106]. Improper coordination between Polβ and FEN1 could lead to a small amount of strand displacement with the formation of a small hairpin in the displaced flap. FEN1 “alternate cleavage” of a short 5’ flap at the base of the hairpin could produce a ligatable nick that after ligation results in incorporation of hairpin bases into the “repaired” strand.(b) Alternatively, a nick close to a repeat loop-out formed during transcription or replication may allow loading of MutSβ/MutSα complexes and the diversion of the normal BER process to produce an MMR-dependent expansion. MMR proteins may also act in the LP BER-based expansion processes shown in (a) to stabilize secondary structures formed by the repeats and perhaps to prevent their removal by enzymes like FEN1. XPA, CSB and RAD52 may act in an auxiliary capacity in either of these pathways via the ability to stabilize secondary structures in the case of XPA, to facilitate incision by OGG1 in the case of RAD52 [61] and to increase incision or to facilitate the formation of an optimal chromatin or transcriptional state in the case of CSB (reviewed in [112]). Loops generated at any stage of the expansion process are susceptible to oxidative damage that could produce result in repeated “toxic oxidation cycles” that could result in multiple rounds of BER-mediated expansions [108].
