Abstract
Substitutional RNA editing changes single C nucleotides in higher plant chloroplast transcripts into U residues. To determine the cis-acting sequence elements involved in plastid RNA editing, we constructed a series of chloroplast transformation vectors harboring selected editing sites of the tobacco ndhB transcript in a chimeric context. The constructs were inserted into the tobacco plastid genome by biolistic transformation leading to the production of stable chimeric RNAs. Analysis of RNA editing revealed unexpected differences in the size of the essential cis elements or in their distance from the editing site. Flanking sequences of identical size direct virtually complete editing for one pair of editing sites, partial editing for a second and no editing at all for a third pair of sites. Serial 5' and 3' deletions allowed us to define the cis-acting elements more precisely and to identify a sequence element essential for editing site recognition. In addition, a single nucleotide substitution immediately upstream of an editing position was introduced. This mutation was found drastically and selectively to reduce the editing efficiency of the downstream editing site, demonstrating that position -1 is important for either site recognition or catalysis. Our results indicate that the editing of adjacent sites is likely to be mechanistically coupled. In no case did the presence in the plastome of the additional editing sites have any effect on the editing efficiency of the endogenous ndhB sites, indicating that the availability of site-specific trans-acting factors is not rate limiting.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araya A., Domec C., Begu D., Litvak S. An in vitro system for the editing of ATP synthase subunit 9 mRNA using wheat mitochondrial extracts. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1040–1044. doi: 10.1073/pnas.89.3.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B., Lüke W., Hunsmann G. Improvement of PCR amplified DNA sequencing with the aid of detergents. Nucleic Acids Res. 1990 Mar 11;18(5):1309–1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
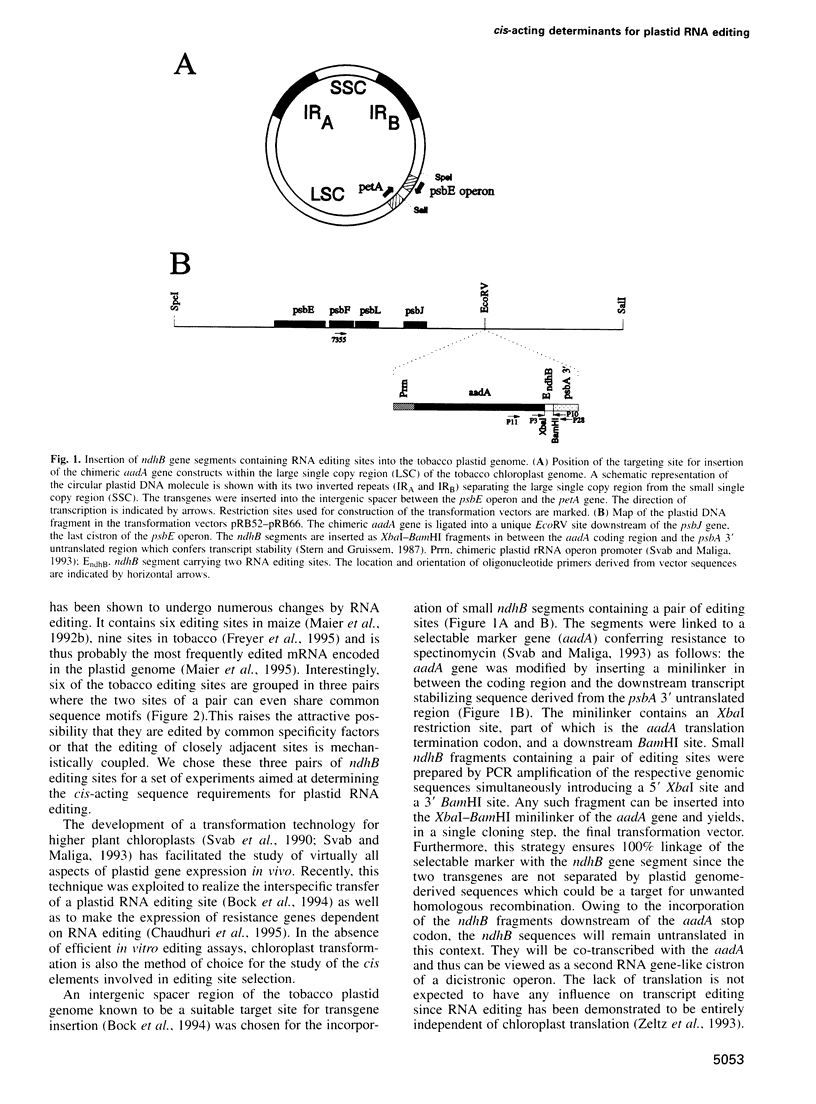
- Bock R., Kössel H., Maliga P. Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J. 1994 Oct 3;13(19):4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L. RNA editing: exploring one mode with apolipoprotein B mRNA. Bioessays. 1993 Jan;15(1):33–41. doi: 10.1002/bies.950150106. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S., Carrer H., Maliga P. Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J. 1995 Jun 15;14(12):2951–2957. doi: 10.1002/j.1460-2075.1995.tb07295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S., Gray M. W. RNA editing in plant mitochondria. Nature. 1989 Oct 19;341(6243):662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Freyer R., Hoch B., Neckermann K., Maier R. M., Kössel H. RNA editing in maize chloroplasts is a processing step independent of splicing and cleavage to monocistronic mRNAs. Plant J. 1993 Oct;4(4):621–629. doi: 10.1046/j.1365-313x.1993.04040621.x. [DOI] [PubMed] [Google Scholar]
- Freyer R., López C., Maier R. M., Martín M., Sabater B., Kössel H. Editing of the chloroplast ndhB encoded transcript shows divergence between closely related members of the grass family (Poaceae). Plant Mol Biol. 1995 Nov;29(4):679–684. doi: 10.1007/BF00041158. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989 Oct 19;341(6243):660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Weil J. H., Grienenberger J. M. Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res. 1990 Jul 11;18(13):3771–3776. doi: 10.1093/nar/18.13.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R., Wissinger B., Schuster W., Brennicke A. RNA editing in plant mitochondria. Science. 1989 Dec 22;246(4937):1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- Hoch B., Maier R. M., Appel K., Igloi G. L., Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991 Sep 12;353(6340):178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- Kanevski I., Maliga P. Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1969–1973. doi: 10.1073/pnas.91.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Igloi G. L., Metzlaff M., Hagemann R., Kössel H. RNA editing in tobacco chloroplasts leads to the formation of a translatable psbL mRNA by a C to U substitution within the initiation codon. EMBO J. 1992 Mar;11(3):1099–1103. doi: 10.1002/j.1460-2075.1992.tb05149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. M., Hoch B., Zeltz P., Kössel H. Internal editing of the maize chloroplast ndhA transcript restores codons for conserved amino acids. Plant Cell. 1992 May;4(5):609–616. doi: 10.1105/tpc.4.5.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. M., Neckermann K., Hoch B., Akhmedov N. B., Kössel H. Identification of editing positions in the ndhB transcript from maize chloroplasts reveals sequence similarities between editing sites of chloroplasts and plant mitochondria. Nucleic Acids Res. 1992 Dec 11;20(23):6189–6194. doi: 10.1093/nar/20.23.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. M., Neckermann K., Igloi G. L., Kössel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 1995 Sep 1;251(5):614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- Matsubayashi T., Wakasugi T., Shinozaki K., Yamaguchi-Shinozaki K., Zaita N., Hidaka T., Meng B. Y., Ohto C., Tanaka M., Kato A. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol Gen Genet. 1987 Dec;210(3):385–393. doi: 10.1007/BF00327187. [DOI] [PubMed] [Google Scholar]
- Ruf S., Zeltz P., Kössel H. Complete RNA editing of unspliced and dicistronic transcripts of the intron-containing reading frame IRF170 from maize chloroplasts. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2295–2299. doi: 10.1073/pnas.91.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Hiesel R., Wissinger B., Brennicke A. RNA editing in the cytochrome b locus of the higher plant Oenothera berteriana includes a U-to-C transition. Mol Cell Biol. 1990 May;10(5):2428–2431. doi: 10.1128/mcb.10.5.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Svab Z., Hajdukiewicz P., Maliga P. Stable transformation of plastids in higher plants. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z., Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Schuster W. Evidence for a site-specific cytidine deamination reaction involved in C to U RNA editing of plant mitochondria. J Biol Chem. 1995 Aug 4;270(31):18227–18233. doi: 10.1074/jbc.270.31.18227. [DOI] [PubMed] [Google Scholar]
- Zeltz P., Hess W. R., Neckermann K., Börner T., Kössel H. Editing of the chloroplast rpoB transcript is independent of chloroplast translation and shows different patterns in barley and maize. EMBO J. 1993 Nov;12(11):4291–4296. doi: 10.1002/j.1460-2075.1993.tb06113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]