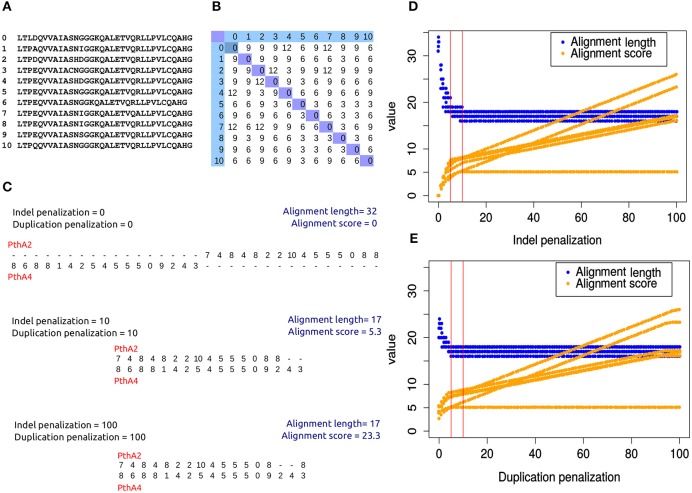
Figure 2.
Indel and duplication penalization for DisTAL. The effect of different penalization values was assessed by generating alignments for 4 TAL effectors from strain IAPAR 306 of Xanthomonas citri pv citri. (A–C) Alignments between PthA2 and PthA4 are shown as an example. (A) Unique TAL repeats found in PthA2 and PthA4. (B) Scoring matrix between repeats as calculated by DisTAL using the Needleman-Wunsch algorithm. (C) Visualization of alignments of the coded sequences of PthA2 and PthA4. (D) Variation in alignment length and score for TAL effectors from Xcc IAPAR 306 using Distal with different indel penalization values. Duplication penalization was kept as 100, each point represents a pairwise alignment between two TAL effectors, red lines indicate range between 5 and 10. (E) Variation in alignment length and score for TAL effectors from Xcc IAPAR 306 using Distal with different duplication penalization values. Indel penalization was kept as 100. Alignment scores were multiplied by 10 for scaling.