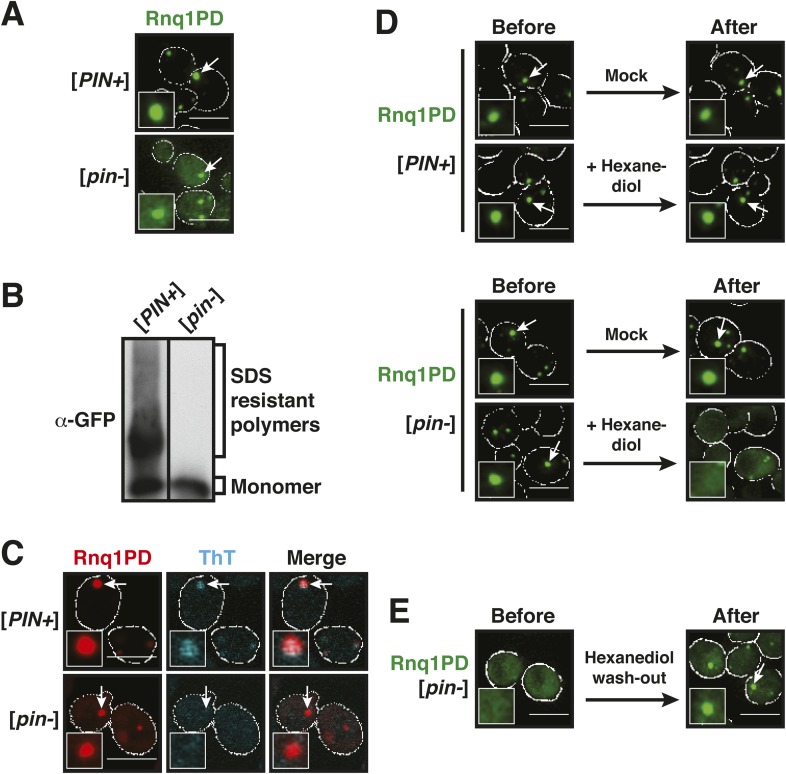
Figure 1. Prion-like domains can access two distinct aggregated states, only one of which is amyloid-like.
(A) Fluorescence microscopy of yeast cells expressing sfGFP-tagged Rnq1PD in [PIN+] and [pin−] cells. White lines indicate the cell boundaries. Scale bars: 5 µm. Also see related Figure 1—figure supplements 1–3. (B) Semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE) of [PIN+] and [pin−] cells containing a plasmid for expression of Rnq1PD-sfGFP. SDS-resistant amyloid polymers show slower migration in comparison to SDS-soluble monomers. Proteins were detected by immunoblotting with a GFP-specific antibody. (C) Thioflavin T (ThT) staining of [PIN+] and [pin−] cells expressing Rnq1PD-mCherry from a plasmid. Note that only amyloid-like assemblies can be stained with ThT. (D) 1,6-hexanediol treatment specifically disrupts non-amyloid Rnq1PD assemblies and not amyloids. Fluorescence time-lapse microscopy of [PIN+] (top panel) and [pin−] cells (bottom panel) expressing Rnq1PD-sfGFP. Time points are before treatment (Before) and 38 min after treatment with 10% 1,6-hexanediol (After). In the control condition (Mock) only media was added. Cells were permeabilized with 10 µg/ ml digitonin. See corresponding Video 1. (E) Cells expressing Rnq1PD-sfGFP were treated with 10% 1,6-hexanediol and digitonin for 1 hr to dissolve non-amyloid Rnq1PD assemblies (Before). Hexanediol was washed out and replaced with normal growth media (After), and the cells were observed with fluorescence microscopy. Also see corresponding Video 2.
Figure 1—figure supplement 1. Lsm4 has a prion-like C-terminal domain (underlined) that is enriched for asparagines (N) and glutamines (Q) and contains hydrophobic residues (L, V, I, M, F).

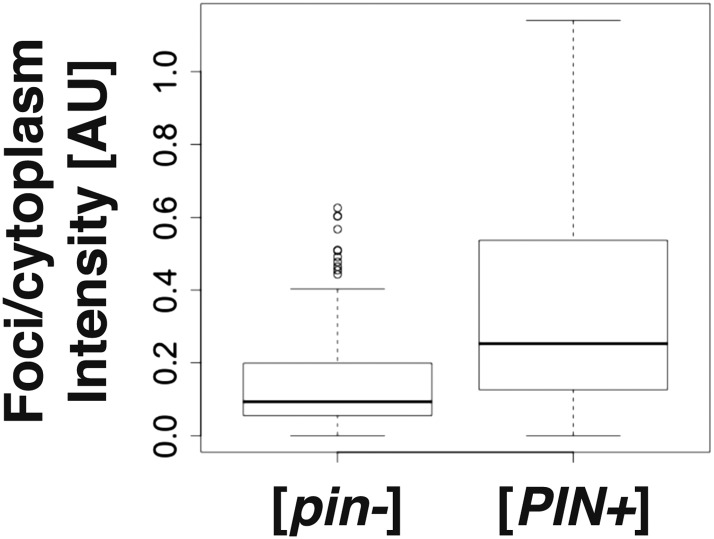
Figure 1—figure supplement 2. Rnq1PD shows a different aggregation pattern in [pin−] and [PIN+] cells.
Figure 1—figure supplement 3. The amino acid sequence of the prion domain of Rnq1 resembles that of FG repeat-containing low-complexity domains of nucleoporins (hydrophobic aromatic residues in an asparagine- and glutamine-rich polar sequence background).