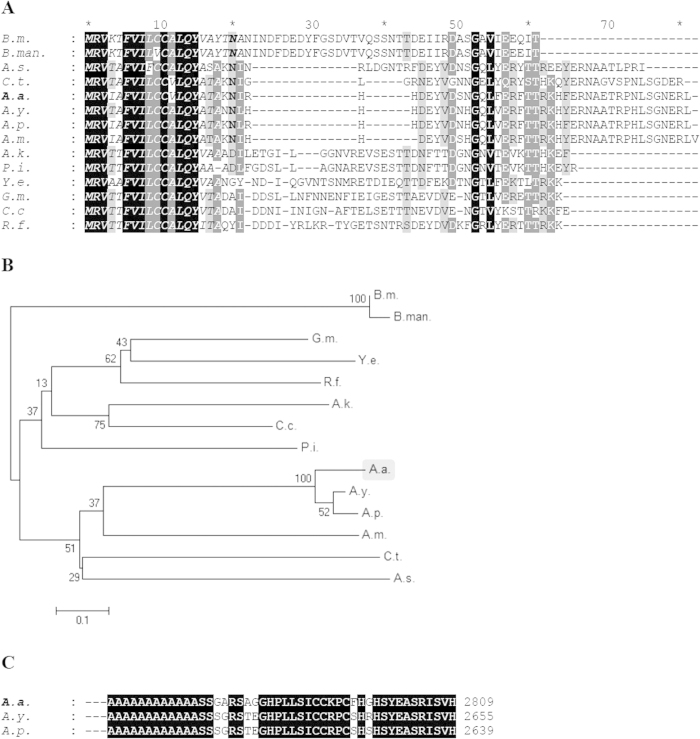
Figure 6. Conservation of terminal non-repetitive sequences of H-fibroin proteins.
(A) the N-terminal sequences from H-fibroin of B. mori (B.m., AF226688), B. mandarina (B.man., X03973), Actias selene (A.s., GU212855), Cricula trifenestrata (C.t., JF264729), A. assama (A.a., KJ862544), A. yamamai (A.y., AB542805), A. pernyi (A.p., AF083334), A. mylitta (A.m., AY136274), Anagasta kuehniella (A.k., AY253534), Plodia interpunctella (P.i., AY253533), Yponomeuta evonymellus (Y.e., AB195979), G. mellonella (G.m., AH009792), Corcyra cephalonica (C.c., GQ901977), Rhodinia fugax (R.f., AB437258) are aligned to show conservation. Highest level of similarity is represented with black background and lesser ones with gray. Signal peptide sequences at N-terminal side are italicized. (B) Unrooted bootstrap consensus phylogenetic tree was generated from the amino acid multiple sequence alignment ‘A’, with bootstrap support values indicated at branch points. The bar represents the number of amino acid replacements per site and AaFhc highlighted with a gray background. (C) conservation at the non-repetitive C-terminal sequences of Antheraea H-fibroins.