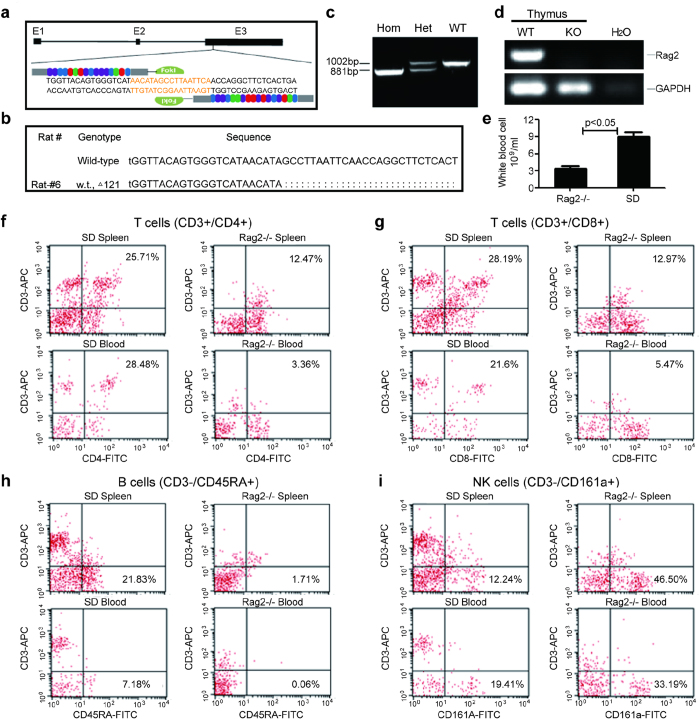
Figure 1. Generation and characterization of Rag2-/- rats.
(a) Schematic representation of Rag2 gene deletion targeting strategy. Exon 3 of Rag2 gene was targeted with the TALEN genome editing technology. (b) Sequencing of the TALEN targeting site identified a 121bp deletion resulting in a frame shift of the Rag2 gene. (c) Genotyping analysis of Rag2-/- heterozygous and homozygous rats using specific primers to amplify Rag2 gene from the tail DNA. WT band, 1002bp; knockout band 881bp. (d) Detection of Rag2 mRNA expression in the thymus by RT-PCR analysis. The Rag2 primer amplifed a 461bp band only in wildtype rat thymus, while GAPDH primers were used as controls and amplifed a band in both wildtype and Rag2-/- rats. (e) Analysis of peripheral white blood cell counts in Rag2-/- and SD rats (n = 10). (f–i) The presence of various lymphocyte populations was examined by flow cytometry in the peripheral blood and spleen of SD and Rag2-/- rat. The T cell compartment was analyzed by simultaneously staining either with APC-CD3 and FITC-CD4 (f) or APC-CD3 and FITC-CD8 (g). To analyze B cell compartment, cells were stained with APC-CD3 and FITC-CD45RA (h). The NK cells were revealed with double staining of APC-CD3 and FITC-CD161a (i). The percentages of cells positive for a given phenotype are indicated by the numbers.